+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10570 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Tail tube of native GTA particle computed with C3 symmetry | ||||||||||||||||||||||||
 Map data Map data | tail tube with tape-measure protein of native GTA particle computed with C3 symmetry | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
| Function / homology | Gene transfer agent, major tail protein / Phage major tail protein TP901-1 / Phage tail tube protein / Phage major tail protein, TP901-1 family Function and homology information Function and homology information | ||||||||||||||||||||||||
| Biological species |  Rhodobacter capsulatus (bacteria) Rhodobacter capsulatus (bacteria) | ||||||||||||||||||||||||
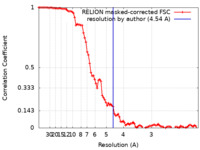
| Method | single particle reconstruction / cryo EM / Resolution: 4.54 Å | ||||||||||||||||||||||||
 Authors Authors | Bardy P / Fuzik T / Hrebik D / Pantucek R / Beatty JT / Plevka P | ||||||||||||||||||||||||
| Funding support |  Czech Republic, 7 items Czech Republic, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Structure and mechanism of DNA delivery of a gene transfer agent. Authors: Pavol Bárdy / Tibor Füzik / Dominik Hrebík / Roman Pantůček / J Thomas Beatty / Pavel Plevka /   Abstract: Alphaproteobacteria, which are the most abundant microorganisms of temperate oceans, produce phage-like particles called gene transfer agents (GTAs) that mediate lateral gene exchange. However, the ...Alphaproteobacteria, which are the most abundant microorganisms of temperate oceans, produce phage-like particles called gene transfer agents (GTAs) that mediate lateral gene exchange. However, the mechanism by which GTAs deliver DNA into cells is unknown. Here we present the structure of the GTA of Rhodobacter capsulatus (RcGTA) and describe the conformational changes required for its DNA ejection. The structure of RcGTA resembles that of a tailed phage, but it has an oblate head shortened in the direction of the tail axis, which limits its packaging capacity to less than 4,500 base pairs of linear double-stranded DNA. The tail channel of RcGTA contains a trimer of proteins that possess features of both tape measure proteins of long-tailed phages from the family Siphoviridae and tail needle proteins of short-tailed phages from the family Podoviridae. The opening of a constriction within the RcGTA baseplate enables the ejection of DNA into bacterial periplasm. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10570.map.gz emd_10570.map.gz | 8.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10570-v30.xml emd-10570-v30.xml emd-10570.xml emd-10570.xml | 16.6 KB 16.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10570_fsc.xml emd_10570_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_10570.png emd_10570.png | 98.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10570 http://ftp.pdbj.org/pub/emdb/structures/EMD-10570 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10570 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10570 | HTTPS FTP |
-Related structure data
| Related structure data |  6tb9C  6tbaC  6te8C  6te9C  6teaC  6tebC  6tehC  6to8C  6toaC  6tsuC  6tsvC  6tswC  6tuiC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10570.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10570.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | tail tube with tape-measure protein of native GTA particle computed with C3 symmetry | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.063 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Rhodobacter capsulatus DE442 gene transfer agent tail tube
| Entire | Name: Rhodobacter capsulatus DE442 gene transfer agent tail tube |
|---|---|
| Components |
|
-Supramolecule #1: Rhodobacter capsulatus DE442 gene transfer agent tail tube
| Supramolecule | Name: Rhodobacter capsulatus DE442 gene transfer agent tail tube type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: tubular structure interconnecting head to baseplate (host recognition device) composed out of 5 disks of tail tube hexamer. Inside tape-measure protein trimer is located. |
|---|---|
| Source (natural) | Organism:  Rhodobacter capsulatus (bacteria) Rhodobacter capsulatus (bacteria) |
| Molecular weight | Theoretical: 500 KDa |
-Macromolecule #1: Tail tube protein Rcc01691
| Macromolecule | Name: Tail tube protein Rcc01691 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Sequence | String: MAAQNGKDLL IKLDLTGSGQ FETIAGLRAT RISFNAETVD VTSLESQGGW RELLGGAGVR SASISGAGVF KDADTDERAR QIFFDGEVPE FQVIIPDFGI VQGPFMITSI DYAGSHNGEA SYELAMASAG ALSFTAI |
-Macromolecule #2: Tape-measure protein Rcc01694
| Macromolecule | Name: Tape-measure protein Rcc01694 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Sequence | String: MVEVDGLDGL GRQAAELERA LGGAEGVAAS FSDELGRMRE SLTYTGREVG TLSSSFGRSL RRAFDGVVFD GMKLSDALKT LAEGMSQAAY SVAMKPVQEA VGGALASTVN GLLGSVFGFA QGGAFSQGRV MPFAKGGVVS SPTSFPMRGA TGLMGEAGPE AILPLARAAD ...String: MVEVDGLDGL GRQAAELERA LGGAEGVAAS FSDELGRMRE SLTYTGREVG TLSSSFGRSL RRAFDGVVFD GMKLSDALKT LAEGMSQAAY SVAMKPVQEA VGGALASTVN GLLGSVFGFA QGGAFSQGRV MPFAKGGVVS SPTSFPMRGA TGLMGEAGPE AILPLARAAD GRLGVQAGGG RVVNVVMNVT TPDAAGFARS QGQIAAQVNR RLARGSRNA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 20 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.8 Component:
Details: G-buffer, doi: 10.1016/0003-9861(77)90508-2 | ||||||||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 11.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: OTHER | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 3114 / Average exposure time: 1.0 sec. / Average electron dose: 42.75 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: -3.0 µm / Nominal defocus min: -1.0 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)