+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10571 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Baseplate of empty GTA particle computed with C3 symmetry | ||||||||||||||||||||||||
 Map data Map data | baseplate of empty GTA particle computed with C3 symmetry | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationGTA TIM-barrel-like domain / : / GTA TIM-barrel-like domain / Rcc01698-like, C-terminal / Protein of unknown function DUF2460 / Conserved hypothetical protein 2217 (DUF2460) / Baseplate hub protein, N-terminal attachment domain / Bacteriophage phiJL001, Gp84 / Bacteriophage phiJL001, Gp84, C-terminal / Phage conserved hypothetical protein BR0599 ...GTA TIM-barrel-like domain / : / GTA TIM-barrel-like domain / Rcc01698-like, C-terminal / Protein of unknown function DUF2460 / Conserved hypothetical protein 2217 (DUF2460) / Baseplate hub protein, N-terminal attachment domain / Bacteriophage phiJL001, Gp84 / Bacteriophage phiJL001, Gp84, C-terminal / Phage conserved hypothetical protein BR0599 / Tip attachment protein J / Putative phage tail protein / Glycoside hydrolase superfamily Similarity search - Domain/homology | ||||||||||||||||||||||||
| Biological species |  Rhodobacter capsulatus DE442 (bacteria) Rhodobacter capsulatus DE442 (bacteria) | ||||||||||||||||||||||||
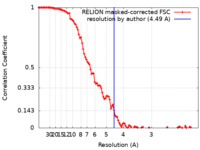
| Method | single particle reconstruction / cryo EM / Resolution: 4.49 Å | ||||||||||||||||||||||||
 Authors Authors | Bardy P / Fuzik T / Hrebik D / Pantucek R / Beatty JT / Plevka P | ||||||||||||||||||||||||
| Funding support |  Czech Republic, 7 items Czech Republic, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Structure and mechanism of DNA delivery of a gene transfer agent. Authors: Pavol Bárdy / Tibor Füzik / Dominik Hrebík / Roman Pantůček / J Thomas Beatty / Pavel Plevka /   Abstract: Alphaproteobacteria, which are the most abundant microorganisms of temperate oceans, produce phage-like particles called gene transfer agents (GTAs) that mediate lateral gene exchange. However, the ...Alphaproteobacteria, which are the most abundant microorganisms of temperate oceans, produce phage-like particles called gene transfer agents (GTAs) that mediate lateral gene exchange. However, the mechanism by which GTAs deliver DNA into cells is unknown. Here we present the structure of the GTA of Rhodobacter capsulatus (RcGTA) and describe the conformational changes required for its DNA ejection. The structure of RcGTA resembles that of a tailed phage, but it has an oblate head shortened in the direction of the tail axis, which limits its packaging capacity to less than 4,500 base pairs of linear double-stranded DNA. The tail channel of RcGTA contains a trimer of proteins that possess features of both tape measure proteins of long-tailed phages from the family Siphoviridae and tail needle proteins of short-tailed phages from the family Podoviridae. The opening of a constriction within the RcGTA baseplate enables the ejection of DNA into bacterial periplasm. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10571.map.gz emd_10571.map.gz | 12.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10571-v30.xml emd-10571-v30.xml emd-10571.xml emd-10571.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10571_fsc.xml emd_10571_fsc.xml | 10.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_10571.png emd_10571.png | 138.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10571 http://ftp.pdbj.org/pub/emdb/structures/EMD-10571 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10571 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10571 | HTTPS FTP |
-Related structure data
| Related structure data |  6tb9C  6tbaC  6te8C  6te9C  6teaC  6tebC  6tehC  6to8C  6toaC  6tsuC  6tsvC  6tswC  6tuiC C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10571.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10571.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | baseplate of empty GTA particle computed with C3 symmetry | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.063 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Rhodobacter capsulatus DE442 gene transfer agent baseplate
| Entire | Name: Rhodobacter capsulatus DE442 gene transfer agent baseplate |
|---|---|
| Components |
|
-Supramolecule #1: Rhodobacter capsulatus DE442 gene transfer agent baseplate
| Supramolecule | Name: Rhodobacter capsulatus DE442 gene transfer agent baseplate type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: host recognition device present at the tip of the tail, empty particle |
|---|---|
| Source (natural) | Organism:  Rhodobacter capsulatus DE442 (bacteria) Rhodobacter capsulatus DE442 (bacteria) |
| Molecular weight | Theoretical: 1.00 MDa |
-Macromolecule #1: Putative gene transfer agent protein
| Macromolecule | Name: Putative gene transfer agent protein / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Sequence | String: MAYPESLKAH LQGGVTTLAR AWALARADGR VLGFTDHDVV LRFDGISFEP GSGMTAKAVL QGTGLSVDNT ESYGALSSEA ITEADLLAG RYDGAAVTVW LVNWADPAMR AVIFRGHLGE VSRGAGAFTA ELRGLTAALG QEQGRIYHPR CAAVLGDGRC R FDLTKDGY ...String: MAYPESLKAH LQGGVTTLAR AWALARADGR VLGFTDHDVV LRFDGISFEP GSGMTAKAVL QGTGLSVDNT ESYGALSSEA ITEADLLAG RYDGAAVTVW LVNWADPAMR AVIFRGHLGE VSRGAGAFTA ELRGLTAALG QEQGRIYHPR CAAVLGDGRC R FDLTKDGY ALEAALGGVD EAVVLRLAEG AGFEDRWFEK GRLVVLDGAA AGLIGVVKND RLQADGSRLI ELWQRLGANP VA GDRVRIE PGCDKRAGTC RLKFDNFLNF RGFPHIPGED WMVSYPVQSG TNDGGSLFR |
-Macromolecule #2: Putative gene transfer agent protein
| Macromolecule | Name: Putative gene transfer agent protein / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Sequence | String: MATILLSAAG AAIGGGFGGT VLGLSGAVIG RAVGATLGRV IDQRLLGSGS QSVETGRVDR LRLSSASEGE AVGRLWGRMR VAGQVIWAT RFFESASVEK SGKGVPRATV TSYSYSLSLA LALCEGEILR VGRVWADGSE IEVSGLNMRV YRGGEDQLPD P KIAAVEGA ...String: MATILLSAAG AAIGGGFGGT VLGLSGAVIG RAVGATLGRV IDQRLLGSGS QSVETGRVDR LRLSSASEGE AVGRLWGRMR VAGQVIWAT RFFESASVEK SGKGVPRATV TSYSYSLSLA LALCEGEILR VGRVWADGSE IEVSGLNMRV YRGGEDQLPD P KIAAVEGA EAAPAYRGIA YVVLEDLQLA PFGNRVPQFT FEVVRAAQGA LAEAEPDLTR GLRAVALIPG TGEYALATTP VY LATGSGV TATQGVANQN APGGQTDLVA ALERLDEELP NCGAVSLVVS WFGDDLRCGA CDVKPKVASV AEEGANMPWR VAG LERAGA EEVPRLSGQS VYGGTPADAA VIEAIAALRA AGKAVTFYPF ILMAQLAGNG LPDPWNPGSA QPALPWRGRI TLSV APGRA GSPDGTAAAE AQVAGFFGAA SPGDSAIAGG EVVYSGPEEW SMRRFILHYA HLCQLAGGVD AFCIGTEMVA LTQIR GPSN SFPAVAAFRQ LAGEVKAILG PGCKIGYAAD WSEYWGYAPG NGERFFHLDP LWADENIDFI GIDNYLPLSD WRDGAD HAD AGWGSIHALD YLRSNIEGGE YYDWFYAAPE HRAAQIRTPI TDGDHDEPWI WRAKDLRNWW LNDHHERVGG LRSEVAT AW VPQSKPIWFT EMGCAAIDKG TNQPNKFLDP KSSESGLPHH SDGRRDELIQ MQYLRAMTGY WGEAARNPVS AVYGGPML D MSRAHVWAWD ARPWPQFPLN TALWSDGENY ARGHWISGRA VAQPLASVVA EICGAAGITE IDVSGLYGLV RGYTMTGDQ TGRAGLQALM LAYGFEALER DGQLVFRMRD GRVAADLAAA DLALGEGEAV VETVRAAEAE IAGRVRLAYV EAEGDFEVKA VEAVFPDAA AGAAAGSELS LALTRAQAQG IVGRWLAEAR VARDTARFAL PPSRGHLGTG DVVRLDLPEG KRRYRIDRVE Q AGLIQVEA VRVEPGIYAP ADEVEDPASL RPFAAPVPVT AVFLDLPLMK GDEDPVAPHL AVTATPWPGT VAVWSSDEDA GY ALNASLG TRAVIGQTLT PLFRARPGVW DRGAALRVRL ASGALDSATA AKVLNGANAM AIGDGSSENW EVFQFAEAAL VEG KIWDIS LRLRGQLGTD ALMPEVWPEG SVVVALNGAP EQILLPSAAR GLARHYRIGA AVRSYDDPSF VHRIEAFAGA GLRP FSPCH LRAEPGASGW AFRWVRRTRI DGDSWQGYEV PLGETAELYL VRVLEGTAVK REVTVGEASW SYPAALQAAD GIAGA FTLE VAQVSDVYGP GLAARITVGA |
-Macromolecule #3: Putative gene transfer agent protein
| Macromolecule | Name: Putative gene transfer agent protein / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Sequence | String: MAFHEVRFPA NLSFGSVGGP ERRTEIVTLS SGHEERNSPW AHSRRHYDAG VGLRSLDDVE RLIAFFEARG GQLHGFRWKD WADFKSCPA SRAVAHEDQL IGMGDGVTTA FQLVKTYVSG GQSYLRPIVK PVEGTVKLGI AGDHQAEAVN FAVDHATGIV S FNEPPPQG ...String: MAFHEVRFPA NLSFGSVGGP ERRTEIVTLS SGHEERNSPW AHSRRHYDAG VGLRSLDDVE RLIAFFEARG GQLHGFRWKD WADFKSCPA SRAVAHEDQL IGMGDGVTTA FQLVKTYVSG GQSYLRPIVK PVEGTVKLGI AGDHQAEAVN FAVDHATGIV S FNEPPPQG ARVTAGFEFD VPVRFDTDRI AVSVQSFQAG DLPQVPVVEV RI |
-Macromolecule #4: Tail fiber protein Rcc00171
| Macromolecule | Name: Tail fiber protein Rcc00171 / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Sequence | String: MADQTPLLGL PLILPSQAQK HVTHNEALSL LDAIVQLAVL DRVRTAPPAS PQTGDRHIVA PGGAAAWTGQ DGAVALWTGS GWLFAQPQPG WTARDLSDGA LLIFDGSLWG PAPVATDNLP GLGINTTHDT TNRLAVQAAA TLLSHAGAGH QLKINKALPT DTASLLFQTS ...String: MADQTPLLGL PLILPSQAQK HVTHNEALSL LDAIVQLAVL DRVRTAPPAS PQTGDRHIVA PGGAAAWTGQ DGAVALWTGS GWLFAQPQPG WTARDLSDGA LLIFDGSLWG PAPVATDNLP GLGINTTHDT TNRLAVQAAA TLLSHAGAGH QLKINKALPT DTASLLFQTS WSGRAEMGTT GSDSFAIKVS ADGTAWKTAF SFAGATGLAS GLAVQQSRTD VTAGRLMRAD WGYGPGNLLG TVAQSAGVPT GAVLERGTTA NGEYIRFADG TQICFAQRPL GSIIAQGAGS FAAPYRTADV SWTYPKVFSA APVVTCRGVA PVGLTQDRRL CAGGVGDLDA TAATGIHVVR LGGAANADVF KADLIAIGPW V |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 20 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.8 Component:
Details: G-buffer, doi: 10.1016/0003-9861(77)90508-2 | ||||||||||||||||||
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 11.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: OTHER | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 3114 / Average exposure time: 1.0 sec. / Average electron dose: 42.75 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: -3.0 µm / Nominal defocus min: -1.0 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)