[English] 日本語
 Yorodumi
Yorodumi- EMDB-10495: Cryo-EM Structure of NADH reduced form of NAD+-dependent Formate ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10495 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of NADH reduced form of NAD+-dependent Formate Dehydrogenase from Rhodobacter capsulatus | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | molybdoenzyme / formate oxidation / NAD+-dependent / OXIDOREDUCTASE | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationformate dehydrogenase complex / formate metabolic process / formate dehydrogenase / formate dehydrogenase / formate dehydrogenase (NAD+) activity / oxidoreductase complex / molybdopterin cofactor binding / NADH dehydrogenase activity / NADH dehydrogenase (ubiquinone) activity / respiratory electron transport chain ...formate dehydrogenase complex / formate metabolic process / formate dehydrogenase / formate dehydrogenase / formate dehydrogenase (NAD+) activity / oxidoreductase complex / molybdopterin cofactor binding / NADH dehydrogenase activity / NADH dehydrogenase (ubiquinone) activity / respiratory electron transport chain / 2 iron, 2 sulfur cluster binding / FMN binding / 4 iron, 4 sulfur cluster binding / electron transfer activity / oxidoreductase activity / metal ion binding / membrane Similarity search - Function | ||||||||||||
| Biological species |  Rhodobacter capsulatus (bacteria) Rhodobacter capsulatus (bacteria) | ||||||||||||
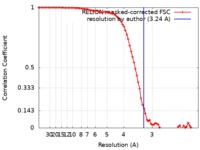
| Method | single particle reconstruction / cryo EM / Resolution: 3.24 Å | ||||||||||||
 Authors Authors | Wendler P / Radon C | ||||||||||||
| Funding support |  Germany, 3 items Germany, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Cryo-EM structures reveal intricate Fe-S cluster arrangement and charging in Rhodobacter capsulatus formate dehydrogenase. Authors: Christin Radon / Gerd Mittelstädt / Benjamin R Duffus / Jörg Bürger / Tobias Hartmann / Thorsten Mielke / Christian Teutloff / Silke Leimkühler / Petra Wendler /   Abstract: Metal-containing formate dehydrogenases (FDH) catalyse the reversible oxidation of formate to carbon dioxide at their molybdenum or tungsten active site. They display a diverse subunit and cofactor ...Metal-containing formate dehydrogenases (FDH) catalyse the reversible oxidation of formate to carbon dioxide at their molybdenum or tungsten active site. They display a diverse subunit and cofactor composition, but structural information on these enzymes is limited. Here we report the cryo-electron microscopic structures of the soluble Rhodobacter capsulatus FDH (RcFDH) as isolated and in the presence of reduced nicotinamide adenine dinucleotide (NADH). RcFDH assembles into a 360 kDa dimer of heterotetramers revealing a putative interconnection of electron pathway chains. In the presence of NADH, the RcFDH structure shows charging of cofactors, indicative of an increased electron load. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10495.map.gz emd_10495.map.gz | 4.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10495-v30.xml emd-10495-v30.xml emd-10495.xml emd-10495.xml | 21 KB 21 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10495_fsc.xml emd_10495_fsc.xml | 8 KB | Display |  FSC data file FSC data file |
| Images |  emd_10495.png emd_10495.png | 93 KB | ||
| Filedesc metadata |  emd-10495.cif.gz emd-10495.cif.gz | 7.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10495 http://ftp.pdbj.org/pub/emdb/structures/EMD-10495 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10495 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10495 | HTTPS FTP |
-Related structure data
| Related structure data |  6tg9MC  6tgaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10495.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10495.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Formate dehydrogenase, NADH reduced
+Supramolecule #1: Formate dehydrogenase, NADH reduced
+Macromolecule #1: Formate dehydrogenase subunit alpha
+Macromolecule #2: Formate dehydrogenase subunit beta
+Macromolecule #3: Formate dehydrogenase subunit gamma
+Macromolecule #4: NAD-dependent formate dehydrogenase subunit delta
+Macromolecule #5: 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,1...
+Macromolecule #6: MOLYBDENUM(VI) ION
+Macromolecule #7: FE2/S2 (INORGANIC) CLUSTER
+Macromolecule #8: IRON/SULFUR CLUSTER
+Macromolecule #9: HYDROSULFURIC ACID
+Macromolecule #10: FLAVIN MONONUCLEOTIDE
+Macromolecule #11: 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||
| Grid | Model: Quantifoil R2/4 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR | ||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy #1
Electron microscopy #1
| Microscopy ID | 1 |
|---|---|
| Microscope | FEI POLARA 300 |
| Image recording | Image recording ID: 1 / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 64.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Electron microscopy #1~
Electron microscopy #1~
| Microscopy ID | 1 |
|---|---|
| Microscope | FEI TITAN KRIOS |
| Image recording | Image recording ID: 2 / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: BACKBONE TRACE | ||||||||
| Output model |  PDB-6tg9: |
 Movie
Movie Controller
Controller























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)