[English] 日本語
 Yorodumi
Yorodumi- EMDB-10470: Cryo-EM structure of Euglena gracilis mitochondrial ATP synthase,... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10470 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Euglena gracilis mitochondrial ATP synthase, rotor, rotational state 1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | mitochondria / ATP synthase / MEMBRANE PROTEIN | |||||||||
| Biological species |  Euglena gracilis (euglena) Euglena gracilis (euglena) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Muhleip A / Amunts A | |||||||||
 Citation Citation |  Journal: Elife / Year: 2019 Journal: Elife / Year: 2019Title: Structure of a mitochondrial ATP synthase with bound native cardiolipin. Authors: Alexander Mühleip / Sarah E McComas / Alexey Amunts /  Abstract: The mitochondrial ATP synthase fuels eukaryotic cells with chemical energy. Here we report the cryo-EM structure of a divergent ATP synthase dimer from mitochondria of , a member of the phylum ...The mitochondrial ATP synthase fuels eukaryotic cells with chemical energy. Here we report the cryo-EM structure of a divergent ATP synthase dimer from mitochondria of , a member of the phylum Euglenozoa that also includes human parasites. It features 29 different subunits, 8 of which are newly identified. The membrane region was determined to 2.8 Å resolution, enabling the identification of 37 associated lipids, including 25 cardiolipins, which provides insight into protein-lipid interactions and their functional roles. The rotor-stator interface comprises four membrane-embedded horizontal helices, including a distinct subunit . The dimer interface is formed entirely by phylum-specific components, and a peripherally associated subcomplex contributes to the membrane curvature. The central and peripheral stalks directly interact with each other. Last, the ATPase inhibitory factor 1 (IF) binds in a mode that is different from human, but conserved in Trypanosomatids. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10470.map.gz emd_10470.map.gz | 288.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10470-v30.xml emd-10470-v30.xml emd-10470.xml emd-10470.xml | 18.4 KB 18.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_10470.png emd_10470.png | 33.8 KB | ||
| Masks |  emd_10470_msk_1.map emd_10470_msk_1.map | 325 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-10470.cif.gz emd-10470.cif.gz | 5.7 KB | ||
| Others |  emd_10470_half_map_1.map.gz emd_10470_half_map_1.map.gz emd_10470_half_map_2.map.gz emd_10470_half_map_2.map.gz | 258.9 MB 260.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10470 http://ftp.pdbj.org/pub/emdb/structures/EMD-10470 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10470 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10470 | HTTPS FTP |
-Related structure data
| Related structure data |  6tdxMC  6tduC  6tdvC  6tdwC  6tdyC  6tdzC  6te0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_10470.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10470.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10470_msk_1.map emd_10470_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
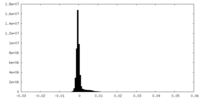
| Density Histograms |
-Half map: #2
| File | emd_10470_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
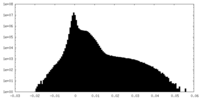
| Density Histograms |
-Half map: #1
| File | emd_10470_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Euglena gracilis mitochondrial ATP synthase dimer
| Entire | Name: Euglena gracilis mitochondrial ATP synthase dimer |
|---|---|
| Components |
|
-Supramolecule #1: Euglena gracilis mitochondrial ATP synthase dimer
| Supramolecule | Name: Euglena gracilis mitochondrial ATP synthase dimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Euglena gracilis (euglena) Euglena gracilis (euglena) |
| Molecular weight | Theoretical: 2 MDa |
-Macromolecule #1: ATP synthase F1 subunit gamma
| Macromolecule | Name: ATP synthase F1 subunit gamma / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Euglena gracilis (euglena) Euglena gracilis (euglena) |
| Molecular weight | Theoretical: 35.110375 KDa |
| Sequence | String: MPGGGTIRFW REKLEGYKKY HQIVKTIKMV TLAKYRQTVV RTRVRDQTLR YTRKALDAKT QDDQEVIEKS ECLLYVPITT NRGSCGALN TNMVRYLQEV ENPKMTIISV GKKALDAMTK VFQDTYRRTI LNDMKQAMSF QFAAYVLEHM NTVPWDRAQI V YNRYHGAA ...String: MPGGGTIRFW REKLEGYKKY HQIVKTIKMV TLAKYRQTVV RTRVRDQTLR YTRKALDAKT QDDQEVIEKS ECLLYVPITT NRGSCGALN TNMVRYLQEV ENPKMTIISV GKKALDAMTK VFQDTYRRTI LNDMKQAMSF QFAAYVLEHM NTVPWDRAQI V YNRYHGAA SQKLAIFNLP KFEDWKQKLE EDSAGDGKIE EDGLLQSLPM KTALGELEET AVEDFYNFHS CLAVLNAVSE NE LSEYAAR IVAVENQLGN ITGLMQLADY TYNKTRKELI TAELLEIIGT MTAMHAGKKV GLKKTEFWK |
-Macromolecule #2: F-type H+-transporting ATPase subunit delta
| Macromolecule | Name: F-type H+-transporting ATPase subunit delta / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Euglena gracilis (euglena) Euglena gracilis (euglena) |
| Molecular weight | Theoretical: 19.559953 KDa |
| Sequence | String: MRASRTLLLS VSRFMRQDPR KFFPDNGFRF FDGPEDSFGD GNIPAQIILT LTRQDEFILK QEPVAAITIR TNEGEMGVLA GHEYTVQQL APGILEVEYE GGKKDQYVIS GGFAHVNDTG VVDINTVEAV PLEEIDHEKL AKALEEARAK SQSPDEAVRI Q GEIALEIF EPLEAALH |
-Macromolecule #3: ATP synthase F1 subunit epsilon
| Macromolecule | Name: ATP synthase F1 subunit epsilon / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Euglena gracilis (euglena) Euglena gracilis (euglena) |
| Molecular weight | Theoretical: 8.751981 KDa |
| Sequence | String: MSWRDAGISY LRYLSIVTRC IHEVQKEGPL LTKNVRFSTI GWKSLYLDHG ATKEYTAIPA ELEKIPENQV AQQHHA |
-Macromolecule #4: ATP synthase subunit c
| Macromolecule | Name: ATP synthase subunit c / type: protein_or_peptide / ID: 4 / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Euglena gracilis (euglena) Euglena gracilis (euglena) |
| Molecular weight | Theoretical: 10.820831 KDa |
| Sequence | String: MQRGSSITKV VRRAALARST RNAAIAYEVT VNGANLIGAG MAASGVGVPA IGVAMCFSSY MLAAARQPNM SAKLLPYCIL GFALSEALA LFTLLIALLE LFVFS |
-Macromolecule #5: ATPEG1
| Macromolecule | Name: ATPEG1 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Euglena gracilis (euglena) Euglena gracilis (euglena) |
| Molecular weight | Theoretical: 19.708709 KDa |
| Sequence | String: MSLAKVWTYA SWIPRGIPKA MANELSSAAA ALAHPEAIAR VAQLESQGKN PYRVARAEFW QMYLACWPYR FRNTVVEWET CKAKVLKGS VDLQDIVDLL YLLAWAYLFW ILGEIYGRGS LYGYRFDGEI HRQEAQNVIL YKEKEAQEMA VVMEKLEKEI Q EWLKTMEQ E |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: 3 seconds blot. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Number real images: 9045 / Average exposure time: 10.0 sec. / Average electron dose: 36.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)