[English] 日本語
 Yorodumi
Yorodumi- EMDB-1015: Cryo-electron microscopy reveals the functional organization of a... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-1015 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
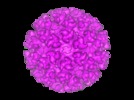
| Title | Cryo-electron microscopy reveals the functional organization of an enveloped virus, Semliki Forest virus. | |||||||||
 Map data Map data | Semliki Forest virus. | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationtogavirin / T=4 icosahedral viral capsid / virion assembly / small molecule binding / host cell endosome / symbiont-mediated suppression of host toll-like receptor signaling pathway / clathrin-dependent endocytosis of virus by host cell / host cell cytoplasm / viral translational frameshifting / serine-type endopeptidase activity ...togavirin / T=4 icosahedral viral capsid / virion assembly / small molecule binding / host cell endosome / symbiont-mediated suppression of host toll-like receptor signaling pathway / clathrin-dependent endocytosis of virus by host cell / host cell cytoplasm / viral translational frameshifting / serine-type endopeptidase activity / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / host cell nucleus / host cell plasma membrane / virion membrane / structural molecule activity / proteolysis / RNA binding / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |   Semliki forest virus Semliki forest virus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 9.0 Å | |||||||||
 Authors Authors | Mancini EJ / Clarke M / Gowen B / Rutten T / Fuller SD | |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2000 Journal: Mol Cell / Year: 2000Title: Cryo-electron microscopy reveals the functional organization of an enveloped virus, Semliki Forest virus. Authors: E J Mancini / M Clarke / B E Gowen / T Rutten / S D Fuller /  Abstract: Semliki Forest virus serves as a paradigm for membrane fusion and assembly. Our icosahedral reconstruction combined 5276 particle images from 48 cryo-electron micrographs and determined the virion ...Semliki Forest virus serves as a paradigm for membrane fusion and assembly. Our icosahedral reconstruction combined 5276 particle images from 48 cryo-electron micrographs and determined the virion structure to 9 A resolution. The improved resolution of this map reveals an N-terminal arm linking capsid subunits and defines the spike-capsid interaction sites. It illustrates the paired helical nature of the transmembrane segments and the elongated structures connecting them to the spike projecting domains. A 10 A diameter density in the fusion protein lines the cavity at the center of the spike. These clearly visible features combine with the variation in order between the layers to provide a framework for understanding the structural changes during the life cycle of an enveloped virus. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_1015.map.gz emd_1015.map.gz | 26.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-1015-v30.xml emd-1015-v30.xml emd-1015.xml emd-1015.xml | 16.6 KB 16.6 KB | Display Display |  EMDB header EMDB header |
| Images |  1015.gif 1015.gif | 37.3 KB | ||
| Others |  emd_1015_additional_1.map emd_1015_additional_1.map emd_1015_additional_2.map emd_1015_additional_2.map emd_1015_additional_3.map emd_1015_additional_3.map | 354.9 KB 354.9 KB 354.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-1015 http://ftp.pdbj.org/pub/emdb/structures/EMD-1015 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1015 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-1015 | HTTPS FTP |
-Related structure data
| Related structure data |  1dylMC  2xfcM M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_1015.map.gz / Format: CCP4 / Size: 50.8 MB / Type: IMAGE STORED AS SIGNED INTEGER (2 BYTES) Download / File: emd_1015.map.gz / Format: CCP4 / Size: 50.8 MB / Type: IMAGE STORED AS SIGNED INTEGER (2 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Semliki Forest virus. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.52 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Supplemental map: emd 1015 additional 1.map
| File | emd_1015_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Supplemental map: emd 1015 additional 2.map
| File | emd_1015_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Supplemental map: emd 1015 additional 3.map
| File | emd_1015_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Semliki Forest Virus
| Entire | Name:   Semliki Forest Virus Semliki Forest Virus |
|---|---|
| Components |
|
-Supramolecule #1000: Semliki Forest Virus
| Supramolecule | Name: Semliki Forest Virus / type: sample / ID: 1000 / Oligomeric state: T=4 membraneous particle / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 50 MDa / Method: theoretical |
-Supramolecule #1: Semliki forest virus
| Supramolecule | Name: Semliki forest virus / type: virus / ID: 1 / Name.synonym: SFV / NCBI-ID: 11033 / Sci species name: Semliki forest virus / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No / Syn species name: SFV |
|---|---|
| Host (natural) | Organism: baby hamster kidney 21 cells (unknown) / synonym: VERTEBRATES |
| Virus shell | Shell ID: 1 / Name: envelope / T number (triangulation number): 4 |
| Virus shell | Shell ID: 2 / Name: nucleocapsid / T number (triangulation number): 4 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: Tris (10mM) NaCl (100 mM) ph 7.4 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 37 K / Instrument: HOMEMADE PLUNGER Details: Vitrification instrument: EMBL plunger with warm humid air spray. plunging at ambient temperature and humidity Method: Blot for 2 sec Graticule grids were used to maintain flatness |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM200FEG/ST |
|---|---|
| Temperature | Average: 105 K |
| Date | Jan 1, 1995 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: ZEISS SCAI / Digitization - Sampling interval: 7 µm / Number real images: 48 / Average electron dose: 8 e/Å2 / Camera length: 44 / Od range: 1 / Bits/pixel: 8 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 7.628 µm / Nominal defocus min: 0.975 µm / Nominal magnification: 50000 |
| Sample stage | Specimen holder: eucentric / Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| CTF correction | Details: ctf multiplication and summation of normalized reconstructions |
|---|---|
| Final reconstruction | Applied symmetry - Point group: I (icosahedral) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 9.0 Å / Resolution method: OTHER / Software - Name: EMBL-ICOS Details: final maps were calculated by making a normalized sum of seperate ctf multiplied maps: Baker, T. S., Olson, N. H., and Fuller, S. D. (1999). Adding the third dimension to virus life cycles: ...Details: final maps were calculated by making a normalized sum of seperate ctf multiplied maps: Baker, T. S., Olson, N. H., and Fuller, S. D. (1999). Adding the third dimension to virus life cycles: Three-Dimensional Reconstruction of Icosahedral Viruses from Cryo-Electron Micrographs. Microbiology and Molecular Biology Reviews 63, 862-922. Fuller, S. D., Butcher, S. J., Cheng, R. H., and Baker, T. S. (1996). Three-dimensional reconstruction of icosahedral particles-the uncommon line. J Struct Biol 116, 48-55. Mancini, E. J., Clarke, M., Gowen, B., Rutten, T., and Fuller, S. D. (2000). Cryo-electron microscopy reveals the functional organization of an enveloped virus, Semliki Forest virus. Molecular Cell 5, 255-266. Mancini, E. J., de Haas, F., and Fuller, S. D. (1997). High-resolution icosahedral reconstruction: fulfilling the promise of cryo-electron microscopy. Structure 5, 741-750. Number images used: 6000 |
| Final angle assignment | Details: sufficient to give maximum inverse eigenvalue of 0.1 |
-Atomic model buiding 1
| Initial model | PDB ID: Chain - #0 - Chain ID: A / Chain - #1 - Chain ID: B / Chain - #2 - Chain ID: C / Chain - #3 - Chain ID: D |
|---|---|
| Software | Name: emfit (Cheng et al 1995) |
| Details | Protocol: rigid body. The capsid protein used for the rigid body refinement was PDB entry 1VCQ |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: R factor and clashes |
| Output model |  PDB-1dyl:  PDB-2xfc: |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)













































