[English] 日本語
 Yorodumi
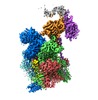
Yorodumi- EMDB-10110: Cryo-EM structure of human oligosaccharyltransferase complex OST-A -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10110 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human oligosaccharyltransferase complex OST-A | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | N-glycosylation / Oligosaccharyltransferase / OSTA / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationoligosaccharyltransferase complex binding / oligosaccharyltransferase complex A / oligosaccharyltransferase complex B / Asparagine N-linked glycosylation / dolichyl-diphosphooligosaccharide-protein glycotransferase / dolichyl-diphosphooligosaccharide-protein glycotransferase activity / oligosaccharyltransferase complex / : / : / : ...oligosaccharyltransferase complex binding / oligosaccharyltransferase complex A / oligosaccharyltransferase complex B / Asparagine N-linked glycosylation / dolichyl-diphosphooligosaccharide-protein glycotransferase / dolichyl-diphosphooligosaccharide-protein glycotransferase activity / oligosaccharyltransferase complex / : / : / : / protein N-linked glycosylation / epithelial cell apoptotic process / azurophil granule membrane / Advanced glycosylation endproduct receptor signaling / blastocyst development / SRP-dependent cotranslational protein targeting to membrane / rough endoplasmic reticulum / response to cytokine / post-translational protein modification / response to endoplasmic reticulum stress / T cell activation / protein modification process / enzyme activator activity / regulation of protein stability / melanosome / transferase activity / Maturation of spike protein / protein-macromolecule adaptor activity / nuclear body / inflammatory response / intracellular membrane-bounded organelle / apoptotic process / Neutrophil degranulation / endoplasmic reticulum membrane / negative regulation of apoptotic process / endoplasmic reticulum / RNA binding / metal ion binding / membrane / plasma membrane / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
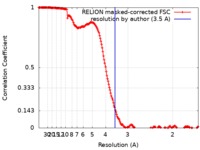
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||
 Authors Authors | Ramirez AS / Kowal J | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2019 Journal: Science / Year: 2019Title: Cryo-electron microscopy structures of human oligosaccharyltransferase complexes OST-A and OST-B. Authors: Ana S Ramírez / Julia Kowal / Kaspar P Locher /  Abstract: Oligosaccharyltransferase (OST) catalyzes the transfer of a high-mannose glycan onto secretory proteins in the endoplasmic reticulum. Mammals express two distinct OST complexes that act in a ...Oligosaccharyltransferase (OST) catalyzes the transfer of a high-mannose glycan onto secretory proteins in the endoplasmic reticulum. Mammals express two distinct OST complexes that act in a cotranslational (OST-A) or posttranslocational (OST-B) manner. Here, we present high-resolution cryo-electron microscopy structures of human OST-A and OST-B. Although they have similar overall architectures, structural differences in the catalytic subunits STT3A and STT3B facilitate contacts to distinct OST subunits, DC2 in OST-A and MAGT1 in OST-B. In OST-A, interactions with TMEM258 and STT3A allow ribophorin-I to form a four-helix bundle that can bind to a translating ribosome, whereas the equivalent region is disordered in OST-B. We observed an acceptor peptide and dolichylphosphate bound to STT3B, but only dolichylphosphate in STT3A, suggesting distinct affinities of the two OST complexes for protein substrates. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10110.map.gz emd_10110.map.gz | 201.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10110-v30.xml emd-10110-v30.xml emd-10110.xml emd-10110.xml | 29.4 KB 29.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10110_fsc.xml emd_10110_fsc.xml | 17.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_10110.png emd_10110.png | 143.2 KB | ||
| Filedesc metadata |  emd-10110.cif.gz emd-10110.cif.gz | 8.6 KB | ||
| Others |  emd_10110_additional_1.map.gz emd_10110_additional_1.map.gz emd_10110_additional_2.map.gz emd_10110_additional_2.map.gz emd_10110_additional_3.map.gz emd_10110_additional_3.map.gz | 183.9 MB 201 MB 201.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10110 http://ftp.pdbj.org/pub/emdb/structures/EMD-10110 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10110 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10110 | HTTPS FTP |
-Related structure data
| Related structure data |  6s7oMC  6s7tC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10110.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10110.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
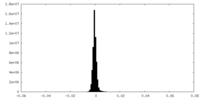
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Local resolution filtered map from RELION
| File | emd_10110_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local resolution filtered map from RELION | ||||||||||||
| Projections & Slices |
| ||||||||||||
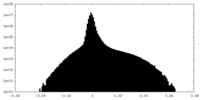
| Density Histograms |
-Additional map: #1
| File | emd_10110_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #2
| File | emd_10110_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : Human oligosaccharyltransferase complex OST-A
+Supramolecule #1: Human oligosaccharyltransferase complex OST-A
+Supramolecule #2: Human oligosaccharyltransferase complex
+Supramolecule #3: Oligosaccharyltransferase complex subunit OSTC
+Macromolecule #1: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #2: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #3: Transmembrane protein 258
+Macromolecule #4: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #5: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #6: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase su...
+Macromolecule #7: Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48...
+Macromolecule #8: Oligosaccharyltransferase complex subunit OSTC
+Macromolecule #12: (2~{S},3~{R},4~{R},5~{S},6~{S})-2-(hydroxymethyl)-6-[(1~{S},2~{R}...
+Macromolecule #13: (4R,7R)-4-hydroxy-N,N,N-trimethyl-4,9-dioxo-7-[(undecanoyloxy)met...
+Macromolecule #14: MAGNESIUM ION
+Macromolecule #15: [(3~{R},6~{Z},10~{Z},14~{Z},18~{Z})-3,7,11,15,19,23-hexamethyltet...
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 3 / Number real images: 6035 / Average exposure time: 8.0 sec. / Average electron dose: 68.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)