+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10027 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mitochondrial 28S ribosome with IF3 (masked refined IF3) | |||||||||
 Map data Map data | Local resolution filtered, masked sharpened map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
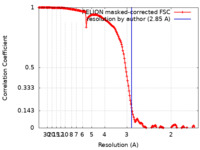
| Method | single particle reconstruction / cryo EM / Resolution: 2.85 Å | |||||||||
 Authors Authors | Itoh Y / Khawaja A / Rorbach J / Amunts A | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Distinct pre-initiation steps in human mitochondrial translation. Authors: Anas Khawaja / Yuzuru Itoh / Cristina Remes / Henrik Spåhr / Olessya Yukhnovets / Henning Höfig / Alexey Amunts / Joanna Rorbach /   Abstract: Translation initiation in human mitochondria relies upon specialized mitoribosomes and initiation factors, mtIF2 and mtIF3, which have diverged from their bacterial counterparts. Here we report two ...Translation initiation in human mitochondria relies upon specialized mitoribosomes and initiation factors, mtIF2 and mtIF3, which have diverged from their bacterial counterparts. Here we report two distinct mitochondrial pre-initiation assembly steps involving those factors. Single-particle cryo-EM revealed that in the first step, interactions between mitochondria-specific protein mS37 and mtIF3 keep the small mitoribosomal subunit in a conformation favorable for a subsequent accommodation of mtIF2 in the second step. Combination with fluorescence cross-correlation spectroscopy analyses suggests that mtIF3 promotes complex assembly without mRNA or initiator tRNA binding, where exclusion is achieved by the N-terminal and C-terminal domains of mtIF3. Finally, the association of large mitoribosomal subunit is required for initiator tRNA and leaderless mRNA recruitment to form a stable initiation complex. These data reveal fundamental aspects of mammalian protein synthesis that are specific to mitochondria. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10027.map.gz emd_10027.map.gz | 3.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10027-v30.xml emd-10027-v30.xml emd-10027.xml emd-10027.xml | 21.7 KB 21.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10027_fsc.xml emd_10027_fsc.xml | 17 KB | Display |  FSC data file FSC data file |
| Images |  emd_10027.png emd_10027.png | 63.5 KB | ||
| Masks |  emd_10027_msk_1.map emd_10027_msk_1.map | 421.9 MB |  Mask map Mask map | |
| Others |  emd_10027_half_map_1.map.gz emd_10027_half_map_1.map.gz emd_10027_half_map_2.map.gz emd_10027_half_map_2.map.gz | 337.4 MB 337.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10027 http://ftp.pdbj.org/pub/emdb/structures/EMD-10027 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10027 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10027 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10027.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10027.map.gz / Format: CCP4 / Size: 7.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local resolution filtered, masked sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10027_msk_1.map emd_10027_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: None
| File | emd_10027_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: None
| File | emd_10027_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | None | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : IF3 bound mitochondrial translation pre-initiation complex
| Entire | Name: IF3 bound mitochondrial translation pre-initiation complex |
|---|---|
| Components |
|
-Supramolecule #1: IF3 bound mitochondrial translation pre-initiation complex
| Supramolecule | Name: IF3 bound mitochondrial translation pre-initiation complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#32 Details: Small subunit of mitochondrial ribosome in complex with mitochondrial IF3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.1 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.7 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 2-20 / Number grids imaged: 1 / Number real images: 13831 / Average exposure time: 4.0 sec. / Average electron dose: 1.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 5.0 µm / Calibrated defocus min: 0.25 µm / Calibrated magnification: 165000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.2 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Target criteria: Cross-correlation coefficient |
 Movie
Movie Controller
Controller




























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)