+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0256 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Natural tetrameric form of human butyrylcholinesterase (BChE) | |||||||||
 Map data Map data | Map after post-processing procedure in RELION 2.1. Contour level was selected based on visualization in Chimera 1.11.2. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
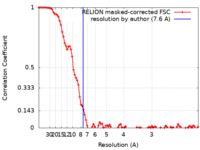
| Method | single particle reconstruction / cryo EM / Resolution: 7.6 Å | |||||||||
 Authors Authors | Baymukhametov TN / Chesnokov YM / Boyko KM / Hons M / Lipkin AV / Lushchekina SV / Konarev PV / Masson P / Vasiliev AL / Popov VO / Kovalchuk MV | |||||||||
| Funding support |  Russian Federation, 2 items Russian Federation, 2 items
| |||||||||
 Citation Citation |  Journal: Biochimie / Year: 2019 Journal: Biochimie / Year: 2019Title: 3D structure of the natural tetrameric form of human butyrylcholinesterase as revealed by cryoEM, SAXS and MD. Authors: Konstantin M Boyko / Timur N Baymukhametov / Yury M Chesnokov / Michael Hons / Sofya V Lushchekina / Petr V Konarev / Alexey V Lipkin / Alexandre L Vasiliev / Patrick Masson / Vladimir O ...Authors: Konstantin M Boyko / Timur N Baymukhametov / Yury M Chesnokov / Michael Hons / Sofya V Lushchekina / Petr V Konarev / Alexey V Lipkin / Alexandre L Vasiliev / Patrick Masson / Vladimir O Popov / Michail V Kovalchuk /   Abstract: Human plasma butyrylcholinesterase (BChE) is an endogenous bioscavenger that hydrolyzes numerous medicamentous and poisonous esters and scavenges potent organophosphorus nerve agents. BChE is thus a ...Human plasma butyrylcholinesterase (BChE) is an endogenous bioscavenger that hydrolyzes numerous medicamentous and poisonous esters and scavenges potent organophosphorus nerve agents. BChE is thus a marker for the diagnosis of OP poisoning. It is also considered a therapeutic target against Alzheimer's disease. Although the X-ray structure of a partially deglycosylated monomer of human BChE was solved 15 years ago, all attempts to determine the 3D structure of the natural full-length glycosylated tetrameric human BChE have been unsuccessful so far. Here, a combination of three complementary structural methods-single-particle cryo-electron microscopy, molecular dynamics and small-angle X-ray scattering-were implemented to elucidate the overall structural and spatial organization of the natural tetrameric human plasma BChE. A 7.6 Å cryoEM map clearly shows the major features of the enzyme: a dimer of dimers with a nonplanar monomer arrangement, in which the interconnecting super helix complex PRAD-(WAT)-peptide C-terminal tail is located in the center of the tetramer, nearly perpendicular to its plane, and is plunged deep between the four subunits. Molecular dynamics simulations allowed optimization of the geometry of the molecule and reconstruction of the structural features invisible in the cryoEM density, i.e., glycan chains and glycan interdimer contact areas, as well as intermonomer disulfide bridges at the C-terminal tail. Finally, SAXS data were used to confirm the consistency of the obtained model with the experimental data. The tetramer organization of BChE is unique in that the four subunits are joined at their C-termini through noncovalent contacts with a short polyproline-rich peptide. This tetramer structure could serve as a model for the design of highly stable glycosylated tetramers. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0256.map.gz emd_0256.map.gz | 5.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0256-v30.xml emd-0256-v30.xml emd-0256.xml emd-0256.xml | 14.4 KB 14.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0256_fsc.xml emd_0256_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_0256.png emd_0256.png | 71 KB | ||
| Masks |  emd_0256_msk_1.map emd_0256_msk_1.map | 64 MB |  Mask map Mask map | |
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0256 http://ftp.pdbj.org/pub/emdb/structures/EMD-0256 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0256 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0256 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0256.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0256.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map after post-processing procedure in RELION 2.1. Contour level was selected based on visualization in Chimera 1.11.2. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.067 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_0256_msk_1.map emd_0256_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Natural tetrameric form of human butyrylcholinesterase
| Entire | Name: Natural tetrameric form of human butyrylcholinesterase |
|---|---|
| Components |
|
-Supramolecule #1: Natural tetrameric form of human butyrylcholinesterase
| Supramolecule | Name: Natural tetrameric form of human butyrylcholinesterase type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 335 KDa |
-Macromolecule #1: Natural tetrameric form of human butyrylcholinesterase
| Macromolecule | Name: Natural tetrameric form of human butyrylcholinesterase type: protein_or_peptide / ID: 1 Details: Glycan chains are attached to the following residues: 17,57,106, 241, 256, 341, 455, 481, 486. Enantiomer: LEVO / EC number: cholinesterase |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MHSKVTIICI RFLFWFLLLC MLIGKSHTED DIIIATKNGK VRGMNLTVFG GTVTAFLGIP YAQPPLGRL RFKKPQSLTK WSDIWNATKY ANSCCQNIDQ SFPGFHGSEM WNPNTDLSED C LYLNVWIP APKPKNATVL IWIYGGGFQT GTSSLHVYDG KFLARVERVI ...String: MHSKVTIICI RFLFWFLLLC MLIGKSHTED DIIIATKNGK VRGMNLTVFG GTVTAFLGIP YAQPPLGRL RFKKPQSLTK WSDIWNATKY ANSCCQNIDQ SFPGFHGSEM WNPNTDLSED C LYLNVWIP APKPKNATVL IWIYGGGFQT GTSSLHVYDG KFLARVERVI VVSMNYRVGA LG FLALPGN PEAPGNMGLF DQQLALQWVQ KNIAAFGGNP KSVTLFGESA GAASVSLHLL SPG SHSLFT RAILQSGSFN APWAVTSLYE ARNRTLNLAK LTGCSRENET EIIKCLRNKD PQEI LLNEA FVVPYGTPLS VNFGPTVDGD FLTDMPDILL ELGQFKKTQI LVGVNKDEGT AFLVY GAPG FSKDNNSIIT RKEFQEGLKI FFPGVSEFGK ESILFHYTDW VDDQRPENYR EALGDV VGD YNFICPALEF TKKFSEWGNN AFFYYFEHRS SKLPWPEWMG VMHGYEIEFV FGLPLER RD NYTKAEEILS RSIVKRWANF AKYGNPNETQ NNSTSWPVFK STEQKYLTLN TESTRIMT K LRAQQCRFWT SFFPKVLEMT GNIDEAEWEW KAGFHRWNNY MMDWKNQFND YTSKKESCVG L |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.026 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV / Details: Blot for 1 second before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-28 / Number grids imaged: 1 / Number real images: 1500 / Average exposure time: 7.0 sec. / Average electron dose: 42.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 46860 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 0.003 µm / Nominal defocus min: 0.001 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)