+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Core Centromere Binding Factor 3 (CBF3) with monomeric Ndc10 | |||||||||
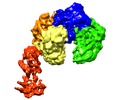
 Map data Map data | Reconstruction of the CBF3CCdeltaN (Core complex of CBF3 comprising the homodimer of Cep3deltaN and the heterodimer of Skp1-Ctf13) in complex with domains 1-2 of Ndc10 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Centromere / CDEIII-binding / LRR domain / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationRAVE complex / Iron uptake and transport / CBF3 complex / regulation of transcription by galactose / : / cellular response to methylmercury / vacuolar proton-transporting V-type ATPase complex assembly / septin ring assembly / mitotic spindle elongation / centromeric DNA binding ...RAVE complex / Iron uptake and transport / CBF3 complex / regulation of transcription by galactose / : / cellular response to methylmercury / vacuolar proton-transporting V-type ATPase complex assembly / septin ring assembly / mitotic spindle elongation / centromeric DNA binding / regulation of exit from mitosis / kinetochore assembly / condensed chromosome, centromeric region / positive regulation of D-glucose transmembrane transport / spindle pole body / exit from mitosis / vacuolar acidification / regulation of metabolic process / protein neddylation / mitotic intra-S DNA damage checkpoint signaling / mitochondrial fusion / silent mating-type cassette heterochromatin formation / DNA binding, bending / SCF ubiquitin ligase complex / FBXL7 down-regulates AURKA during mitotic entry and in early mitosis / SCF-dependent proteasomal ubiquitin-dependent protein catabolic process / mitotic spindle assembly checkpoint signaling / Orc1 removal from chromatin / spindle midzone / Antigen processing: Ubiquitination & Proteasome degradation / DNA replication origin binding / cullin family protein binding / regulation of protein-containing complex assembly / subtelomeric heterochromatin formation / negative regulation of cytoplasmic translation / endomembrane system / regulation of mitotic cell cycle / chromosome segregation / G1/S transition of mitotic cell cycle / kinetochore / G2/M transition of mitotic cell cycle / spindle / mitotic cell cycle / protein-containing complex assembly / ubiquitin-dependent protein catabolic process / DNA-binding transcription factor activity, RNA polymerase II-specific / chromosome, telomeric region / protein ubiquitination / DNA binding / zinc ion binding / identical protein binding / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Zhang WJ / Lukoynova N | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2018 Journal: Cell Rep / Year: 2018Title: Insights into Centromere DNA Bending Revealed by the Cryo-EM Structure of the Core Centromere Binding Factor 3 with Ndc10. Authors: Wenjuan Zhang / Natalya Lukoyanova / Shomon Miah / Jonathan Lucas / Cara K Vaughan /  Abstract: The centromere binding factor 3 (CBF3) complex binds the third centromere DNA element in organisms with point centromeres, such as S. cerevisiae. It is an essential complex for assembly of the ...The centromere binding factor 3 (CBF3) complex binds the third centromere DNA element in organisms with point centromeres, such as S. cerevisiae. It is an essential complex for assembly of the kinetochore in these organisms, as it facilitates genetic centromere specification and allows association of all other kinetochore components. We determined high-resolution structures of the core complex of CBF3 alone and in association with a monomeric construct of Ndc10, using cryoelectron microscopy (cryo-EM). We identify the DNA-binding site of the complex and present a model in which CBF3 induces a tight bend in centromeric DNA, thus facilitating assembly of the centromeric nucleosome. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|---|
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0051.map.gz emd_0051.map.gz | 59.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0051-v30.xml emd-0051-v30.xml emd-0051.xml emd-0051.xml | 19 KB 19 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0051_fsc.xml emd_0051_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_0051.png emd_0051.png | 129.6 KB | ||
| Filedesc metadata |  emd-0051.cif.gz emd-0051.cif.gz | 7.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0051 http://ftp.pdbj.org/pub/emdb/structures/EMD-0051 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0051 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0051 | HTTPS FTP |
-Related structure data
| Related structure data |  6gsaMC  0052C  4241C  6fe8C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_0051.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0051.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of the CBF3CCdeltaN (Core complex of CBF3 comprising the homodimer of Cep3deltaN and the heterodimer of Skp1-Ctf13) in complex with domains 1-2 of Ndc10 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Core CBF3 in complex with Ndc10 D1-2
| Entire | Name: Core CBF3 in complex with Ndc10 D1-2 |
|---|---|
| Components |
|
-Supramolecule #1: Core CBF3 in complex with Ndc10 D1-2
| Supramolecule | Name: Core CBF3 in complex with Ndc10 D1-2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: The truncated CBF3 complex, recombinantly expressed in Saccharomyces cerevisiae. It comprises a Cep3 homodimer, in which the binuclear zinc cluster domains are truncated, full length ...Details: The truncated CBF3 complex, recombinantly expressed in Saccharomyces cerevisiae. It comprises a Cep3 homodimer, in which the binuclear zinc cluster domains are truncated, full length heterodimer of Skp1 and Ctf13, and a monomeric construct Ndc10 comprising domains 1-2. |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 286 KDa |
-Macromolecule #1: Centromere DNA-binding protein complex CBF3 subunit B
| Macromolecule | Name: Centromere DNA-binding protein complex CBF3 subunit B / type: protein_or_peptide / ID: 1 Details: N-terminal polyhistidine purification tagTruncation of the binuclear zinc cluster domain Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 68.454125 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGGSSHHHHH HSSGLVPRGS HMKLITASSS KEYLPDLLLF WQNYEYWITN IGLYKTKQRD LTRTPANLDT DTEECMFWMN YLQKDQSFQ LMNFAMENLG ALYFGSIGDI SELYLRVEQY WDRRADKNHS VDGKYWDALI WSVFTMCIYY MPVEKLAEIF S VYPLHEYL ...String: MGGSSHHHHH HSSGLVPRGS HMKLITASSS KEYLPDLLLF WQNYEYWITN IGLYKTKQRD LTRTPANLDT DTEECMFWMN YLQKDQSFQ LMNFAMENLG ALYFGSIGDI SELYLRVEQY WDRRADKNHS VDGKYWDALI WSVFTMCIYY MPVEKLAEIF S VYPLHEYL GSNKRLNWED GMQLVMCQNF ARCSLFQLKQ CDFMAHPDIR LVQAYLILAT TTFPYDEPLL ANSLLTQCIH TF KNFHVDD FRPLLNDDPV ESIAKVTLGR IFYRLCGCDY LQSGPRKPIA LHTEVSSLLQ HAAYLQDLPN VDVYREENST EVL YWKIIS LDRDLDQYLN KSSKPPLKTL DAIRRELDIF QYKVDSLEED FRSNNSRFQK FIALFQISTV SWKLFKMYLI YYDT ADSLL KVIHYSKVII SLIVNNFHAK SEFFNRHPMV MQTITRVVSF ISFYQIFVES AAVKQLLVDL TELTANLPTI FGSKL DKLV YLTERLSKLK LLWDKVQLLD SGDSFYHPVF KILQNDIKII ELKNDEMFSL IKGLGSLVPL NKLRQESLLE EEDENN TEP SDFRTIVEEF QSEYNISDIL S UniProtKB: Centromere DNA-binding protein complex CBF3 subunit B |
-Macromolecule #2: Suppressor of kinetochore protein 1
| Macromolecule | Name: Suppressor of kinetochore protein 1 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 22.558451 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGVTSNVVLV SGEGERFTVD KKIAERSLLL KNYLNDMHDS NLQNNSDSES DSDSETNHKS KDNNNGDDDD EDDDEIVMPV PNVRSSVLQ KVIEWAEHHR DSNFPDEDDD DSRKSAPVDS WDREFLKVDQ EMLYEIILAA NYLNIKPLLD AGCKVVAEMI R GRSPEEIR ...String: MGVTSNVVLV SGEGERFTVD KKIAERSLLL KNYLNDMHDS NLQNNSDSES DSDSETNHKS KDNNNGDDDD EDDDEIVMPV PNVRSSVLQ KVIEWAEHHR DSNFPDEDDD DSRKSAPVDS WDREFLKVDQ EMLYEIILAA NYLNIKPLLD AGCKVVAEMI R GRSPEEIR RTFNIVNDFT PEEEAAIRRE NEWAEDRGS UniProtKB: Suppressor of kinetochore protein 1 |
-Macromolecule #3: Centromere DNA-binding protein complex CBF3 subunit C
| Macromolecule | Name: Centromere DNA-binding protein complex CBF3 subunit C / type: protein_or_peptide / ID: 3 / Details: C-terminal CBP purification tag / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 60.899961 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGPSFNPVRF LELPIDIRKE VYFHLDGNFC GAHPYPIDIL YKSNDVELPG KPSYKRSKRS KKLLRYMYPV FATYLNIFEY SPQLIEKWL EYAFWLRYDC LVLDCFKVNH LYDGTLIDAL EWTYLDNELR LAYFNKASML EVWYTFKEYK KWVIDSVAFD E LDLLNVSN ...String: MGPSFNPVRF LELPIDIRKE VYFHLDGNFC GAHPYPIDIL YKSNDVELPG KPSYKRSKRS KKLLRYMYPV FATYLNIFEY SPQLIEKWL EYAFWLRYDC LVLDCFKVNH LYDGTLIDAL EWTYLDNELR LAYFNKASML EVWYTFKEYK KWVIDSVAFD E LDLLNVSN IQFNIDNLTP QLVDKCLSIL EQKDLFATIG EVQFGQDEEV GEEKDVDVSG ANSDENSSPS STIKNKKRSA SK RSHSDNG NVGATHNQLT SISVIRTIRS MESMKSLRKI TVRGEKLYEL LINFHGFRDN PGKTISYIVK RRINEIRLSR MNQ ISRTGL ADFTRWDNLQ KLVLSRVAYI DLNSIVFPKN FKSLTMKRVS KIKWWNIEEN ILKELKVDKR TFKSLYIKED DSKF TKFFN LRHTRIKELD KSEINQITYL RCQAIVWLSF RTLNHIKLQN VSEVFNNIIV PRALFDSKRV EIYRCEKISQ VLVIG SRSG SENLYFQGSK RRWKKNFIAV SAANRFKKIS SSGAL UniProtKB: Centromere DNA-binding protein complex CBF3 subunit C |
-Macromolecule #4: Centromere DNA-binding protein complex CBF3 subunit A
| Macromolecule | Name: Centromere DNA-binding protein complex CBF3 subunit A / type: protein_or_peptide / ID: 4 Details: Domains 1-2 of Ndc10 with a non-cleavable C-terminal StrepII tag Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 66.159578 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGRSSILFLL KLMKIMDVQQ QQEAMSSEDR FQELVDSLKP RTAHQYKTYY TKYIQWCQLN QIIPTPEDNS VNSVPYKDLP ISAELIHWF LLDTLITDDK PGEKREETED LDEEEENSFK IATLKKIIGS LNFLSKLCKV HENPNANIDT KYLESVTKLH T HWIDSQKA ...String: MGRSSILFLL KLMKIMDVQQ QQEAMSSEDR FQELVDSLKP RTAHQYKTYY TKYIQWCQLN QIIPTPEDNS VNSVPYKDLP ISAELIHWF LLDTLITDDK PGEKREETED LDEEEENSFK IATLKKIIGS LNFLSKLCKV HENPNANIDT KYLESVTKLH T HWIDSQKA ITTNETNNTN TQVLCPPLLK VSLNLWNPET NHLSEKFFKT CSEKLRFLVD FQLRSYLNLS FEERSKIRFG SL KLGKRDR DAIIYHKVTH SAEKKDTPGH HQLLALLPQD CPFICPQTTL AAYLYLRFYG IPSVSKGDGF PNLNADENGS LLQ DIPILR GKSLTTYPRE ETFSNYYTTV FRYCHLPYKR REYFNKCNLV YPTWDEDTFR TFFNEENHGN WLEQPEAFAF PDKI PFDFK KIMNFKSPYT SYSTNAKKDP FPPPKDLLVQ IFPEIDEYKR HDYEGLSQNS RDFLDLMEVL RERFLSNLPW IYKFF PNHD IFQDPIFGNS DFQSYFNDKT IHSKGSPILS FDILPGFNKI YKNKTNFYSL LIERPSQLTF ASSHNPDTHP WSHPQF EK UniProtKB: Centromere DNA-binding protein complex CBF3 subunit A |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.15 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Grid | Model: Quantifoil, UltrAuFoil, R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK I | ||||||||||||
| Details | The sample is homogeneous and well-dispersed on grids. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 2003 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 47170 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | "Fit in map" function used to place 6FE8 and 4ACO with out further refinement or model building |
|---|---|
| Output model |  PDB-6gsa: |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)