[English] 日本語
 Yorodumi
Yorodumi- EMDB-0052: Masked Core Centromere Binding Factor 3 (CBF3) with monomeric Ndc10 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Masked Core Centromere Binding Factor 3 (CBF3) with monomeric Ndc10 | |||||||||
 Map data Map data | focused refinement of Ndc10D1-2 in CBF3 | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
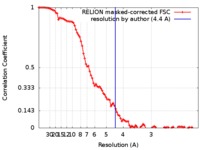
| Method | single particle reconstruction / cryo EM / Resolution: 4.4 Å | |||||||||
 Authors Authors | Zhang WJ / Lukoynova N | |||||||||
 Citation Citation |  Journal: Cell Rep / Year: 2018 Journal: Cell Rep / Year: 2018Title: Insights into Centromere DNA Bending Revealed by the Cryo-EM Structure of the Core Centromere Binding Factor 3 with Ndc10. Authors: Wenjuan Zhang / Natalya Lukoyanova / Shomon Miah / Jonathan Lucas / Cara K Vaughan /  Abstract: The centromere binding factor 3 (CBF3) complex binds the third centromere DNA element in organisms with point centromeres, such as S. cerevisiae. It is an essential complex for assembly of the ...The centromere binding factor 3 (CBF3) complex binds the third centromere DNA element in organisms with point centromeres, such as S. cerevisiae. It is an essential complex for assembly of the kinetochore in these organisms, as it facilitates genetic centromere specification and allows association of all other kinetochore components. We determined high-resolution structures of the core complex of CBF3 alone and in association with a monomeric construct of Ndc10, using cryoelectron microscopy (cryo-EM). We identify the DNA-binding site of the complex and present a model in which CBF3 induces a tight bend in centromeric DNA, thus facilitating assembly of the centromeric nucleosome. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|---|
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0052.map.gz emd_0052.map.gz | 4.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0052-v30.xml emd-0052-v30.xml emd-0052.xml emd-0052.xml | 15.9 KB 15.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0052_fsc.xml emd_0052_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_0052.png emd_0052.png | 162.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0052 http://ftp.pdbj.org/pub/emdb/structures/EMD-0052 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0052 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0052 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0052.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0052.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | focused refinement of Ndc10D1-2 in CBF3 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : Core CBF3 in complex with Ndc10 D1-2
| Entire | Name: Core CBF3 in complex with Ndc10 D1-2 |
|---|---|
| Components |
|
-Supramolecule #1: Core CBF3 in complex with Ndc10 D1-2
| Supramolecule | Name: Core CBF3 in complex with Ndc10 D1-2 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 Details: The truncated CBF3 complex, recombinantly expressed in Saccharomyces cerevisiae. It comprises a Cep3 homodimer, in which the binuclear zinc cluster domains are truncated, full length ...Details: The truncated CBF3 complex, recombinantly expressed in Saccharomyces cerevisiae. It comprises a Cep3 homodimer, in which the binuclear zinc cluster domains are truncated, full length heterodimer of Skp1 and Ctf13, and a monomeric construct Ndc10 comprising domains 1-2. |
|---|---|
| Molecular weight | Theoretical: 286 KDa |
-Supramolecule #2: Cep3 homodimer
| Supramolecule | Name: Cep3 homodimer / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Supramolecule #3: Skp1
| Supramolecule | Name: Skp1 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Supramolecule #4: Ctf13
| Supramolecule | Name: Ctf13 / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Supramolecule #5: Ndc10 comprising domains 1-2
| Supramolecule | Name: Ndc10 comprising domains 1-2 / type: complex / ID: 5 / Parent: 1 / Macromolecule list: #4 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.15 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK I | ||||||||||||
| Details | The sample is homogeneous and well-dispersed on grids. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-40 / Number grids imaged: 1 / Number real images: 2003 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 47170 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
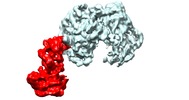
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | "Fit in map" function used to place 6FE8 and 4ACO with out further refinement or model building |
|---|
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)