[English] 日本語
 Yorodumi
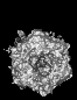
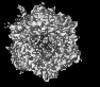
Yorodumi- PDB-9cmo: Cryo-EM model derived from localized reconstruction of Ad657-hexo... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 9cmo | ||||||
|---|---|---|---|---|---|---|---|
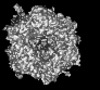
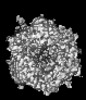
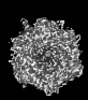
| Title | Cryo-EM model derived from localized reconstruction of Ad657-hexon-FII complex at 4.14A resolution | ||||||
 Components Components |
| ||||||
 Keywords Keywords | VIRAL PROTEIN / Adenovirus / Hexon / Coagulation factor X / Coagulation factor II / Prothrombin / Complex / Interactions / VIRUS | ||||||
| Function / homology |  Function and homology information Function and homology informationT=25 icosahedral viral capsid / microtubule-dependent intracellular transport of viral material towards nucleus / : / thrombospondin receptor activity / Defective factor XII causes hereditary angioedema / thrombin / thrombin-activated receptor signaling pathway / negative regulation of astrocyte differentiation / regulation of blood coagulation / neutrophil-mediated killing of gram-negative bacterium ...T=25 icosahedral viral capsid / microtubule-dependent intracellular transport of viral material towards nucleus / : / thrombospondin receptor activity / Defective factor XII causes hereditary angioedema / thrombin / thrombin-activated receptor signaling pathway / negative regulation of astrocyte differentiation / regulation of blood coagulation / neutrophil-mediated killing of gram-negative bacterium / positive regulation of phospholipase C-activating G protein-coupled receptor signaling pathway / Defective F8 cleavage by thrombin / Platelet Aggregation (Plug Formation) / ligand-gated ion channel signaling pathway / positive regulation of collagen biosynthetic process / negative regulation of platelet activation / negative regulation of blood coagulation / positive regulation of blood coagulation / negative regulation of fibrinolysis / regulation of cytosolic calcium ion concentration / Transport of gamma-carboxylated protein precursors from the endoplasmic reticulum to the Golgi apparatus / Gamma-carboxylation of protein precursors / Common Pathway of Fibrin Clot Formation / Removal of aminoterminal propeptides from gamma-carboxylated proteins / fibrinolysis / Intrinsic Pathway of Fibrin Clot Formation / negative regulation of proteolysis / negative regulation of cytokine production involved in inflammatory response / Peptide ligand-binding receptors / Regulation of Complement cascade / positive regulation of release of sequestered calcium ion into cytosol / acute-phase response / Cell surface interactions at the vascular wall / positive regulation of receptor signaling pathway via JAK-STAT / growth factor activity / lipopolysaccharide binding / positive regulation of insulin secretion / platelet activation / response to wounding / positive regulation of protein localization to nucleus / Golgi lumen / Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) / positive regulation of reactive oxygen species metabolic process / blood coagulation / antimicrobial humoral immune response mediated by antimicrobial peptide / regulation of cell shape / heparin binding / : / host cell / Thrombin signalling through proteinase activated receptors (PARs) / positive regulation of cell growth / blood microparticle / G alpha (q) signalling events / cell surface receptor signaling pathway / positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / receptor ligand activity / endoplasmic reticulum lumen / signaling receptor binding / serine-type endopeptidase activity / positive regulation of cell population proliferation / calcium ion binding / symbiont entry into host cell / host cell nucleus / structural molecule activity / proteolysis / extracellular space / extracellular exosome / extracellular region / plasma membrane Similarity search - Function | ||||||
| Biological species |  Human adenovirus 6 Human adenovirus 6 Homo sapiens (human) Homo sapiens (human) | ||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 4.17 Å | ||||||
 Authors Authors | Reddy, V.S. / Ma, O.X. | ||||||
| Funding support |  United States, 1items United States, 1items
| ||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structure-derived insights from blood factors binding to the surfaces of different adenoviruses. Authors: Haley E Mudrick / Shao-Chia Lu / Janarjan Bhandari / Mary E Barry / Jack R Hemsath / Felix G M Andres / Olivia X Ma / Michael A Barry / Vijay S Reddy /  Abstract: The tropism of adenoviruses (Ads) is significantly influenced by the binding of various blood factors. To investigate differences in their binding, we conducted cryo-EM analysis on complexes of ...The tropism of adenoviruses (Ads) is significantly influenced by the binding of various blood factors. To investigate differences in their binding, we conducted cryo-EM analysis on complexes of several human adenoviruses with human platelet factor-4 (PF4), coagulation factors FII (Prothrombin), and FX. While we observed EM densities for FII and FX bound to all the species-C adenoviruses examined, no densities were seen for PF4, even though PF4 can co-pellet with various Ads. Similar to FX, the γ-carboxyglutamic acid (Gla) domain of FII binds within the surface cavity of hexon trimers. While FII binds equally to species-C Ads: Ad5, Ad6, and Ad657, FX exhibits significantly better binding to Ad5 and Ad657 compared to Ad6. Although only the FX-Gla domain is observed at high-resolution (3.7 Å), the entire FX is visible at low-resolution bound to Ad5 in three equivalent binding modes consistent with the 3-fold symmetric hexon. Only the Gla and kringle-1 domains of FII are visible on all the species-C adenoviruses, where the rigid FII binds in an upright fashion, in contrast to the flexible and bent FX. These data suggest that differential binding of FII and FX may shield certain species-C adenoviruses differently against immune molecules, thereby modulating their tropism. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  9cmo.cif.gz 9cmo.cif.gz | 594.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb9cmo.ent.gz pdb9cmo.ent.gz | 480.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  9cmo.json.gz 9cmo.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/cm/9cmo https://data.pdbj.org/pub/pdb/validation_reports/cm/9cmo ftp://data.pdbj.org/pub/pdb/validation_reports/cm/9cmo ftp://data.pdbj.org/pub/pdb/validation_reports/cm/9cmo | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  45751MC  9cliC  9clnC  9clsC  9cm2C  9cm9C C: citing same article ( M: map data used to model this data |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 108257.406 Da / Num. of mol.: 3 / Source method: isolated from a natural source / Source: (natural)  Human adenovirus 6 / References: UniProt: A0A348FV85 Human adenovirus 6 / References: UniProt: A0A348FV85#2: Protein | | Mass: 70562.930 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)  Homo sapiens (human) / References: UniProt: P00734, thrombin Homo sapiens (human) / References: UniProt: P00734, thrombin#3: Chemical | ChemComp-CA / Has ligand of interest | Y | Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight |
| ||||||||||||||||||||||||
| Source (natural) |
| ||||||||||||||||||||||||
| Details of virus | Empty: NO / Enveloped: NO / Isolate: SPECIES / Type: VIRION | ||||||||||||||||||||||||
| Natural host | Organism: Human adenovirus 6 | ||||||||||||||||||||||||
| Virus shell |
| ||||||||||||||||||||||||
| Buffer solution | pH: 7.2 | ||||||||||||||||||||||||
| Buffer component | Conc.: 20 mM / Name: Hepes | ||||||||||||||||||||||||
| Specimen | Conc.: 2 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES | ||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 81000 X / Calibrated magnification: 81000 X / Nominal defocus max: 4000 nm / Nominal defocus min: 2500 nm / Calibrated defocus min: 1000 nm / Calibrated defocus max: 5000 nm / Cs: 2.7 mm / C2 aperture diameter: 100 µm / Alignment procedure: COMA FREE |
| Specimen holder | Cryogen: NITROGEN / Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Electron dose: 81 e/Å2 / Film or detector model: GATAN K3 BIOCONTINUUM (6k x 4k) / Num. of grids imaged: 3 / Num. of real images: 10000 |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Type: NONE | ||||||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 59000 | ||||||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 4.17 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 4635 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||||||
| Atomic model building | Protocol: FLEXIBLE FIT | ||||||||||||||||||||||||||||||||||||||||
| Atomic model building | PDB-ID: 6B1T Accession code: 6B1T / Source name: PDB / Type: experimental model | ||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller








 PDBj
PDBj














