[English] 日本語
 Yorodumi
Yorodumi- EMDB-45729: Cryo-EM model derived from localized reconstruction of human aden... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 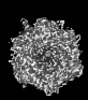 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM model derived from localized reconstruction of human adenovirus 6 (Ad6)-hexon-FII complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Adenovirus / Hexon / Coagulation factor X / Coagulation factor II / Prothrombin / Complex / Interactions / VIRUS / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationT=25 icosahedral viral capsid / microtubule-dependent intracellular transport of viral material towards nucleus / : / thrombospondin receptor activity / Defective factor XII causes hereditary angioedema / thrombin / thrombin-activated receptor signaling pathway / negative regulation of astrocyte differentiation / regulation of blood coagulation / neutrophil-mediated killing of gram-negative bacterium ...T=25 icosahedral viral capsid / microtubule-dependent intracellular transport of viral material towards nucleus / : / thrombospondin receptor activity / Defective factor XII causes hereditary angioedema / thrombin / thrombin-activated receptor signaling pathway / negative regulation of astrocyte differentiation / regulation of blood coagulation / neutrophil-mediated killing of gram-negative bacterium / positive regulation of phospholipase C-activating G protein-coupled receptor signaling pathway / Defective F8 cleavage by thrombin / Platelet Aggregation (Plug Formation) / ligand-gated ion channel signaling pathway / positive regulation of collagen biosynthetic process / negative regulation of platelet activation / negative regulation of blood coagulation / positive regulation of blood coagulation / negative regulation of fibrinolysis / regulation of cytosolic calcium ion concentration / Transport of gamma-carboxylated protein precursors from the endoplasmic reticulum to the Golgi apparatus / Gamma-carboxylation of protein precursors / Common Pathway of Fibrin Clot Formation / Removal of aminoterminal propeptides from gamma-carboxylated proteins / fibrinolysis / Intrinsic Pathway of Fibrin Clot Formation / negative regulation of proteolysis / negative regulation of cytokine production involved in inflammatory response / Peptide ligand-binding receptors / Regulation of Complement cascade / positive regulation of release of sequestered calcium ion into cytosol / acute-phase response / Cell surface interactions at the vascular wall / positive regulation of receptor signaling pathway via JAK-STAT / growth factor activity / lipopolysaccharide binding / positive regulation of insulin secretion / platelet activation / response to wounding / positive regulation of protein localization to nucleus / Golgi lumen / Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) / positive regulation of reactive oxygen species metabolic process / blood coagulation / antimicrobial humoral immune response mediated by antimicrobial peptide / regulation of cell shape / heparin binding / : / host cell / Thrombin signalling through proteinase activated receptors (PARs) / positive regulation of cell growth / blood microparticle / G alpha (q) signalling events / cell surface receptor signaling pathway / positive regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / receptor ligand activity / endoplasmic reticulum lumen / signaling receptor binding / serine-type endopeptidase activity / positive regulation of cell population proliferation / calcium ion binding / symbiont entry into host cell / host cell nucleus / structural molecule activity / proteolysis / extracellular space / extracellular exosome / extracellular region / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Human adenovirus 6 / Human adenovirus 6 /  homo sapiens (human) / homo sapiens (human) /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Reddy VS / Ma OX | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structure-derived insights from blood factors binding to the surfaces of different adenoviruses. Authors: Haley E Mudrick / Shao-Chia Lu / Janarjan Bhandari / Mary E Barry / Jack R Hemsath / Felix G M Andres / Olivia X Ma / Michael A Barry / Vijay S Reddy /  Abstract: The tropism of adenoviruses (Ads) is significantly influenced by the binding of various blood factors. To investigate differences in their binding, we conducted cryo-EM analysis on complexes of ...The tropism of adenoviruses (Ads) is significantly influenced by the binding of various blood factors. To investigate differences in their binding, we conducted cryo-EM analysis on complexes of several human adenoviruses with human platelet factor-4 (PF4), coagulation factors FII (Prothrombin), and FX. While we observed EM densities for FII and FX bound to all the species-C adenoviruses examined, no densities were seen for PF4, even though PF4 can co-pellet with various Ads. Similar to FX, the γ-carboxyglutamic acid (Gla) domain of FII binds within the surface cavity of hexon trimers. While FII binds equally to species-C Ads: Ad5, Ad6, and Ad657, FX exhibits significantly better binding to Ad5 and Ad657 compared to Ad6. Although only the FX-Gla domain is observed at high-resolution (3.7 Å), the entire FX is visible at low-resolution bound to Ad5 in three equivalent binding modes consistent with the 3-fold symmetric hexon. Only the Gla and kringle-1 domains of FII are visible on all the species-C adenoviruses, where the rigid FII binds in an upright fashion, in contrast to the flexible and bent FX. These data suggest that differential binding of FII and FX may shield certain species-C adenoviruses differently against immune molecules, thereby modulating their tropism. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_45729.map.gz emd_45729.map.gz | 768.4 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-45729-v30.xml emd-45729-v30.xml emd-45729.xml emd-45729.xml | 19.8 KB 19.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_45729_fsc.xml emd_45729_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_45729.png emd_45729.png | 51.8 KB | ||
| Filedesc metadata |  emd-45729.cif.gz emd-45729.cif.gz | 7.1 KB | ||
| Others |  emd_45729_half_map_1.map.gz emd_45729_half_map_1.map.gz emd_45729_half_map_2.map.gz emd_45729_half_map_2.map.gz | 22.8 MB 22.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-45729 http://ftp.pdbj.org/pub/emdb/structures/EMD-45729 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45729 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45729 | HTTPS FTP |
-Related structure data
| Related structure data |  9clsMC  9cliC  9clnC  9cm2C  9cm9C  9cmoC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_45729.map.gz / Format: CCP4 / Size: 4.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_45729.map.gz / Format: CCP4 / Size: 4.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.408 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_45729_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_45729_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human adenovirus 6 (Ad6) in complex with Coagulation factor FII
| Entire | Name: Human adenovirus 6 (Ad6) in complex with Coagulation factor FII |
|---|---|
| Components |
|
-Supramolecule #1: Human adenovirus 6 (Ad6) in complex with Coagulation factor FII
| Supramolecule | Name: Human adenovirus 6 (Ad6) in complex with Coagulation factor FII type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: Cryo-EM model derived from localized reconstruction |
|---|---|
| Source (natural) | Organism:  Human adenovirus 6 Human adenovirus 6 |
| Molecular weight | Theoretical: 70 kDa/nm |
-Supramolecule #2: Hexon protein
| Supramolecule | Name: Hexon protein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Human adenovirus 6 Human adenovirus 6 |
-Supramolecule #3: Human Coagulation Factor II
| Supramolecule | Name: Human Coagulation Factor II / type: organelle_or_cellular_component / ID: 3 / Parent: 2 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  homo sapiens (human) homo sapiens (human) |
-Macromolecule #1: Hexon protein
| Macromolecule | Name: Hexon protein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human adenovirus 6 Human adenovirus 6 |
| Molecular weight | Theoretical: 108.635133 KDa |
| Sequence | String: MATPSMMPQW SYMHISGQDA SEYLSPGLVQ FARATETYFS LNNKFRNPTV APTHDVTTDR SQRLTLRFIP VDREDTAYSY KARFTLAVG DNRVLDMAST YFDIRGVLDR GPTFKPYSGT AYNALAPKGA PNSCEWEQNE TAQVDAQELD EEENEANEAQ A REQEQAKK ...String: MATPSMMPQW SYMHISGQDA SEYLSPGLVQ FARATETYFS LNNKFRNPTV APTHDVTTDR SQRLTLRFIP VDREDTAYSY KARFTLAVG DNRVLDMAST YFDIRGVLDR GPTFKPYSGT AYNALAPKGA PNSCEWEQNE TAQVDAQELD EEENEANEAQ A REQEQAKK THVYAQAPLS GIKITKEGLQ IGTADATVAG AGKEIFADKT FQPEPQVGES QWNEADATAA GGRVLKKTTP MK PCYGSYA RPTNSNGGQG VMVEQNGKLE SQVEMQFFST STNATNEVNN IQPTVVLYSE DVNMETPDTH LSYKPKMGDK NAK VMLGQQ AMPNRPNYIA FRDNFIGLMY YNSTGNMGVL AGQASQLNAV VDLQDRNTEL SYQLLLDSIG DRTRYFSMWN QAVD SYDPD VRIIENHGTE DELPNYCFPL GGIGITDTFQ AVKTTAANGD QGNTTWQKDS TFAERNEIGV GNNFAMEINL NANLW RNFL YSNIALYLPD KLKYNPTNVE ISDNPNTYDY MNKRVVAPGL VDCYINLGAR WSLEYMDNVN PFNHHRNAGL RYRSML LGN GRYVPFHIQV PQKFFAIKNL LLLPGSYTYE WNFRKDVNMV LQSSLGNDLR VDGASIKFDS ICLYATFFPM AHNTAST LE AMLRNDTNDQ SFNDYLSAAN MLYPIPANAT NVPISIPSRN WAAFRGWAFT RLKTKETPSL GSGYDPYYTY SGSIPYLD G TFYLNHTFKK VAITFDSSVS WPGNDRLLTP NEFEIKRSVD GEGYNVAQCN MTKDWFLVQM LANYNIGYQG FYIPESYKD RMYSFFRNFQ PMSRQVVDDT KYKDYQQVGI IHQHNNSGFV GYLAPTMREG QAYPANVPYP LIGKTAVDSI TQKKFLCDRT LWRIPFSSN FMSMGALTDL GQNLLYANSA HALDMTFEVD PMDEPTLLYV LFEVFDVVRV HQPHRGVIET VYLRTPFSAG N ATT UniProtKB: Hexon protein |
-Macromolecule #2: Prothrombin
| Macromolecule | Name: Prothrombin / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: thrombin |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 70.56293 KDa |
| Sequence | String: MAHVRGLQLP GCLALAALCS LVHSQHVFLA PQQARSLLQR VRRANTFL(CGU)(CGU) VRKGNL(CGU)R(CGU)C V (CGU)(CGU)TCSY(CGU)(CGU) AF(CGU)AL(CGU)SSTA TDVFWAKYTA CETARTPRDK LAACLEGNCA EGLGTNYR G HVNITRSGIE ...String: MAHVRGLQLP GCLALAALCS LVHSQHVFLA PQQARSLLQR VRRANTFL(CGU)(CGU) VRKGNL(CGU)R(CGU)C V (CGU)(CGU)TCSY(CGU)(CGU) AF(CGU)AL(CGU)SSTA TDVFWAKYTA CETARTPRDK LAACLEGNCA EGLGTNYR G HVNITRSGIE CQLWRSRYPH KPEINSTTHP GADLQENFCR NPDSSTTGPW CYTTDPTVRR QECSIPVCGQ DQVTVAMTP RSEGSSVNLS PPLEQCVPDR GQQYQGRLAV TTHGLPCLAW ASAQAKALSK HQDFNSAVQL VENFCRNPDG DEEGVWCYVA GKPGDFGYC DLNYCEEAVE EETGDGLDED SDRAIEGRTA TSEYQTFFNP RTFGSGEADC GLRPLFEKKS LEDKTERELL E SYIDGRIV EGSDAEIGMS PWQVMLFRKS PQELLCGASL ISDRWVLTAA HCLLYPPWDK NFTENDLLVR IGKHSRTRYE RN IEKISML EKIYIHPRYN WRENLDRDIA LMKLKKPVAF SDYIHPVCLP DRETAASLLQ AGYKGRVTGW GNLKETWTAN VGK GQPSVL QVVNLPIVER PVCKDSTRIR ITDNMFCAGY KPDEGKRGDA CEGDSGGPFV MKSPFNNRWY QMGIVSWGEG CDRD GKYGF YTHVFRLKKW IQKVIDQFGE UniProtKB: Prothrombin |
-Macromolecule #3: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 3 / Number of copies: 7 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.2 / Component - Concentration: 20.0 mM / Component - Name: Hepes |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOCONTINUUM (6k x 4k) / Number grids imaged: 3 / Number real images: 10000 / Average electron dose: 81.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 5.0 µm / Calibrated defocus min: 1.0 µm / Calibrated magnification: 81000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 2.5 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





















 Y (Sec.)
Y (Sec.) X (Row.)
X (Row.) Z (Col.)
Z (Col.)






































