[English] 日本語
 Yorodumi
Yorodumi- EMDB-45744: Cryo-EM model derived from localized reconstruction of Ad657-hexo... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM model derived from localized reconstruction of Ad657-hexon-FX complex at 3.86A resolution | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Adenovirus / Hexon / Coagulation factor X / Coagulation factor II / Prothrombin / Complex / Interactions / VIRUS / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationT=25 icosahedral viral capsid / coagulation factor Xa / microtubule-dependent intracellular transport of viral material towards nucleus / positive regulation of TOR signaling / Golgi lumen / blood coagulation / host cell / endoplasmic reticulum lumen / serine-type endopeptidase activity / calcium ion binding ...T=25 icosahedral viral capsid / coagulation factor Xa / microtubule-dependent intracellular transport of viral material towards nucleus / positive regulation of TOR signaling / Golgi lumen / blood coagulation / host cell / endoplasmic reticulum lumen / serine-type endopeptidase activity / calcium ion binding / symbiont entry into host cell / host cell nucleus / structural molecule activity / proteolysis / extracellular region Similarity search - Function | |||||||||
| Biological species |  Human adenovirus 6 / Human adenovirus 6 /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Reddy VS / Ma OX | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structure-derived insights from blood factors binding to the surfaces of different adenoviruses. Authors: Haley E Mudrick / Shao-Chia Lu / Janarjan Bhandari / Mary E Barry / Jack R Hemsath / Felix G M Andres / Olivia X Ma / Michael A Barry / Vijay S Reddy /  Abstract: The tropism of adenoviruses (Ads) is significantly influenced by the binding of various blood factors. To investigate differences in their binding, we conducted cryo-EM analysis on complexes of ...The tropism of adenoviruses (Ads) is significantly influenced by the binding of various blood factors. To investigate differences in their binding, we conducted cryo-EM analysis on complexes of several human adenoviruses with human platelet factor-4 (PF4), coagulation factors FII (Prothrombin), and FX. While we observed EM densities for FII and FX bound to all the species-C adenoviruses examined, no densities were seen for PF4, even though PF4 can co-pellet with various Ads. Similar to FX, the γ-carboxyglutamic acid (Gla) domain of FII binds within the surface cavity of hexon trimers. While FII binds equally to species-C Ads: Ad5, Ad6, and Ad657, FX exhibits significantly better binding to Ad5 and Ad657 compared to Ad6. Although only the FX-Gla domain is observed at high-resolution (3.7 Å), the entire FX is visible at low-resolution bound to Ad5 in three equivalent binding modes consistent with the 3-fold symmetric hexon. Only the Gla and kringle-1 domains of FII are visible on all the species-C adenoviruses, where the rigid FII binds in an upright fashion, in contrast to the flexible and bent FX. These data suggest that differential binding of FII and FX may shield certain species-C adenoviruses differently against immune molecules, thereby modulating their tropism. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_45744.map.gz emd_45744.map.gz | 714.7 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-45744-v30.xml emd-45744-v30.xml emd-45744.xml emd-45744.xml | 19.8 KB 19.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_45744_fsc.xml emd_45744_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_45744.png emd_45744.png | 55.7 KB | ||
| Filedesc metadata |  emd-45744.cif.gz emd-45744.cif.gz | 7 KB | ||
| Others |  emd_45744_half_map_1.map.gz emd_45744_half_map_1.map.gz emd_45744_half_map_2.map.gz emd_45744_half_map_2.map.gz | 23.2 MB 23.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-45744 http://ftp.pdbj.org/pub/emdb/structures/EMD-45744 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45744 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45744 | HTTPS FTP |
-Related structure data
| Related structure data |  9cm9MC  9cliC  9clnC  9clsC  9cm2C  9cmoC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_45744.map.gz / Format: CCP4 / Size: 4.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_45744.map.gz / Format: CCP4 / Size: 4.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
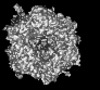
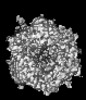
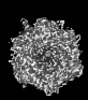
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.408 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_45744_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_45744_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human adenovirus 657 in complex with Coagulation factor FX
| Entire | Name: Human adenovirus 657 in complex with Coagulation factor FX |
|---|---|
| Components |
|
-Supramolecule #1: Human adenovirus 657 in complex with Coagulation factor FX
| Supramolecule | Name: Human adenovirus 657 in complex with Coagulation factor FX type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Human adenovirus 6 Human adenovirus 6 |
| Molecular weight | Theoretical: 55 kDa/nm |
-Supramolecule #2: Hexon protein
| Supramolecule | Name: Hexon protein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Human adenovirus 6 Human adenovirus 6 |
-Supramolecule #3: Human Coagulation Factor X
| Supramolecule | Name: Human Coagulation Factor X / type: organelle_or_cellular_component / ID: 3 / Parent: 2 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Hexon protein
| Macromolecule | Name: Hexon protein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human adenovirus 6 Human adenovirus 6 |
| Molecular weight | Theoretical: 108.257406 KDa |
| Sequence | String: MATPSMMPQW SYMHISGQDA SEYLSPGLVQ FARATETYFS LNNKFRNPTV APTHDVTTDR SQRLTLRFIP VDREDTAYSY KARFTLAVG DNRVLDMAST YFDIRGVLDR GPTFKPYSGT AYNALAPKGA PNSCEWDEDD TQVQVAAEDD QDDDEEEEQL P QQRNGKKT ...String: MATPSMMPQW SYMHISGQDA SEYLSPGLVQ FARATETYFS LNNKFRNPTV APTHDVTTDR SQRLTLRFIP VDREDTAYSY KARFTLAVG DNRVLDMAST YFDIRGVLDR GPTFKPYSGT AYNALAPKGA PNSCEWDEDD TQVQVAAEDD QDDDEEEEQL P QQRNGKKT HVYAQAPFAG EAINKNGLQI GTNGAATEGN KEIYADKTYQ PEPQIGESQW NEAESSVAGG RVLKKTTPMK PC YGSYARP TNSNGGQGVM VEQNGKLESQ VEMQFFSTSV NAMNEANAIQ PKLLLYSEDV NMETPDTHLS YKPGKSDDNS KAM LGQQSM PNRPNYIAFR DNFIGLMYYN STGNMGVLAG QASQLNAVVD LQDRNTELSY QLLLDSIGDR TRYFSMWNQA VDSY DPDVR IIENHGTEDE LPNYCFPLGG IGVTDTYQAI KATNGNGGAT TWAQDNTFAE RNEIGVGNNF AMEINLNANL WRNFL YSNI ALYLPDKLKY NPTNVEISDN PNTYDYMNKR VVAPGLVDCY INLGARWSLD YMDNVNPFNH HRNAGLRYRS MLLGNG RYV PFHIQVPQKF FAIKNLLLLP GSYTYEWNFR KDVNMVLQSS LGNDLRVDGA SIKFDSICLY ATFFPMAHNT ASTLEAM LR NDTNDQSFND YLSAANMLYP IPANATNVPI SIPSRNWAAF RGWAFTRLKT KETPSLGSGY DPYYTYSGSI PYLDGTFY L NHTFKKVAIT FDSSVSWPGN DRLLTPNEFE IKRSVDGEGY NVAQCNMTKD WFLVQMLANY NIGYQGFYIP ESYKDRMYS FFRNFQPMSR QVVDDTKYKD YQQVGIIHQH NNSGFVGYLA PTMREGQAYP ANVPYPLIGK TAVDSITQKK FLCDRTLWRI PFSSNFMSM GALTDLGQNL LYANSAHALD MTFEVDPMDE PTLLYVLFEV FDVVRVHQPH RGVIETVYLR TPFSAGNATT UniProtKB: Hexon protein |
-Macromolecule #2: Coagulation factor X
| Macromolecule | Name: Coagulation factor X / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: coagulation factor Xa |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 55.286738 KDa |
| Sequence | String: MGRPLHLVLL SASLAGLLLL GESLFIRREQ ANNILARVTR ANSFL(CGU)(CGU)MKK GHL(CGU)R(CGU)CM(CGU) (CGU)TCSY(CGU)(CGU)AR(CGU) VF(CGU)DSDKTN(CGU) FWNKYKDGDQ CETSPCQNQG KCKDGLGEYT CTCLEG FEG KNCELFTRKL ...String: MGRPLHLVLL SASLAGLLLL GESLFIRREQ ANNILARVTR ANSFL(CGU)(CGU)MKK GHL(CGU)R(CGU)CM(CGU) (CGU)TCSY(CGU)(CGU)AR(CGU) VF(CGU)DSDKTN(CGU) FWNKYKDGDQ CETSPCQNQG KCKDGLGEYT CTCLEG FEG KNCELFTRKL CSLDNGDCDQ FCHEEQNSVV CSCARGYTLA DNGKACIPTG PYPCGKQTLE RRKRSVAQAT SSSGEAP DS ITWKPYDAAD LDPTENPFDL LDFNQTQPER GDNNLTRIVG GQECKDGECP WQALLINEEN EGFCGGTILS EFYILTAA H CLYQAKRFKV RVGDRNTEQE EGGEAVHEVE VVIKHNRFTK ETYDFDIAVL RLKTPITFRM NVAPACLPER DWAESTLMT QKTGIVSGFG RTHEKGRQST RLKMLEVPYV DRNSCKLSSS FIITQNMFCA GYDTKQEDAC QGDSGGPHVT RFKDTYFVTG IVSWGEGCA RKGKYGIYTK VTAFLKWIDR SMKTRGLPKA KSHAPEVITS SPLK UniProtKB: Coagulation factor X |
-Macromolecule #3: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 3 / Number of copies: 7 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.2 / Component - Concentration: 20.0 mM / Component - Name: Hepes |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOCONTINUUM (6k x 4k) / Number grids imaged: 3 / Number real images: 10000 / Average electron dose: 81.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 5.0 µm / Calibrated defocus min: 1.0 µm / Calibrated magnification: 81000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 2.5 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Y (Sec.)
Y (Sec.) X (Row.)
X (Row.) Z (Col.)
Z (Col.)






































