+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-3341 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Atomic cryoEM structure of Hsp90/Cdc37/Cdk4 complex | |||||||||
 マップデータ マップデータ | Reconstruction of Hsp90:Cdc37:Cdk4 complex. Part of series of maps, the highest resolution map being EMD-3337. This is a different subclass from the same particles as in EMD-3337, having well defined Cdk4 N-Lobe. Other relevant maps are EMD-3338, EMD-3339, EMD-3340. | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | Hsp90 / Cdc37 / Cdk4 / chaperone / kinase / unfolding | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報cyclin D3-CDK4 complex / cyclin D1-CDK4 complex / cyclin D2-CDK4 complex / Evasion of Oncogene Induced Senescence Due to Defective p16INK4A binding to CDK4 / Evasion of Oxidative Stress Induced Senescence Due to Defective p16INK4A binding to CDK4 / cellular response to ionomycin / citrulline metabolic process / regulation of transcription initiation by RNA polymerase II / Drug-mediated inhibition of CDK4/CDK6 activity / Evasion of Oncogene Induced Senescence Due to Defective p16INK4A binding to CDK4 and CDK6 ...cyclin D3-CDK4 complex / cyclin D1-CDK4 complex / cyclin D2-CDK4 complex / Evasion of Oncogene Induced Senescence Due to Defective p16INK4A binding to CDK4 / Evasion of Oxidative Stress Induced Senescence Due to Defective p16INK4A binding to CDK4 / cellular response to ionomycin / citrulline metabolic process / regulation of transcription initiation by RNA polymerase II / Drug-mediated inhibition of CDK4/CDK6 activity / Evasion of Oncogene Induced Senescence Due to Defective p16INK4A binding to CDK4 and CDK6 / Evasion of Oxidative Stress Induced Senescence Due to Defective p16INK4A binding to CDK4 and CDK6 / regulation of type II interferon-mediated signaling pathway / regulation of type B pancreatic cell proliferation / hypoxia-inducible factor-asparagine dioxygenase / : / [protein]-asparagine 3-dioxygenase activity / HSP90-CDC37 chaperone complex / receptor ligand inhibitor activity / very long-chain fatty acid metabolic process / peptidyl-histidine dioxygenase activity / peptidyl-aspartic acid 3-dioxygenase activity / negative regulation of proteasomal protein catabolic process / Aryl hydrocarbon receptor signalling / : / Cellular response to hypoxia / aryl hydrocarbon receptor complex / dynein axonemal particle / carboxylic acid binding / histone methyltransferase binding / positive regulation of vasculogenesis / Transcriptional regulation by RUNX2 / cellular response to phorbol 13-acetate 12-myristate / mitochondrial genome maintenance / ankyrin repeat binding / ATP-dependent protein binding / positive regulation of protein localization to cell surface / protein kinase regulator activity / Notch binding / protein folding chaperone complex / oxygen sensor activity / cyclin-dependent protein serine/threonine kinase regulator activity / telomerase holoenzyme complex assembly / post-transcriptional regulation of gene expression / Respiratory syncytial virus genome replication / Uptake and function of diphtheria toxin / regulation of cyclin-dependent protein serine/threonine kinase activity / Drug-mediated inhibition of ERBB2 signaling / Resistance of ERBB2 KD mutants to trastuzumab / Resistance of ERBB2 KD mutants to sapitinib / Resistance of ERBB2 KD mutants to tesevatinib / Resistance of ERBB2 KD mutants to neratinib / Resistance of ERBB2 KD mutants to osimertinib / Resistance of ERBB2 KD mutants to afatinib / Resistance of ERBB2 KD mutants to AEE788 / Resistance of ERBB2 KD mutants to lapatinib / Drug resistance in ERBB2 TMD/JMD mutants / TPR domain binding / negative regulation of Notch signaling pathway / PTK6 Regulates Cell Cycle / Assembly and release of respiratory syncytial virus (RSV) virions / positive regulation of transforming growth factor beta receptor signaling pathway / dendritic growth cone / Defective binding of RB1 mutants to E2F1,(E2F2, E2F3) / regulation of type I interferon-mediated signaling pathway / The NLRP3 inflammasome / : / Sema3A PAK dependent Axon repulsion / regulation of protein ubiquitination / telomere maintenance via telomerase / HSF1-dependent transactivation / response to unfolded protein / NF-kappaB binding / chaperone-mediated protein complex assembly / HSF1 activation / bicellular tight junction / Attenuation phase / positive regulation of myoblast differentiation / cyclin-dependent kinase / RHOBTB2 GTPase cycle / protein targeting / cyclin-dependent protein serine/threonine kinase activity / cellular response to interleukin-4 / Purinergic signaling in leishmaniasis infection / axonal growth cone / DNA polymerase binding / cyclin-dependent protein kinase holoenzyme complex / supramolecular fiber organization / chaperone-mediated protein folding / Signaling by ERBB2 / heat shock protein binding / negative regulation of proteasomal ubiquitin-dependent protein catabolic process / regulation of G2/M transition of mitotic cell cycle / protein folding chaperone / HSP90 chaperone cycle for steroid hormone receptors (SHR) in the presence of ligand / positive regulation of G2/M transition of mitotic cell cycle / nitric-oxide synthase regulator activity / cyclin binding / Constitutive Signaling by Overexpressed ERBB2 / ESR-mediated signaling / Ubiquitin-dependent degradation of Cyclin D 類似検索 - 分子機能 | |||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||
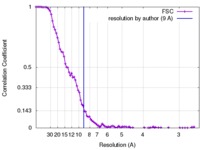
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 9.0 Å | |||||||||
 データ登録者 データ登録者 | Verba KA / Wang RYR / Arakawa A / Liu Y / Shirouzu M / Yokoyama S / Agard DA | |||||||||
 引用 引用 |  ジャーナル: Science / 年: 2016 ジャーナル: Science / 年: 2016タイトル: Atomic structure of Hsp90-Cdc37-Cdk4 reveals that Hsp90 traps and stabilizes an unfolded kinase. 著者: Kliment A Verba / Ray Yu-Ruei Wang / Akihiko Arakawa / Yanxin Liu / Mikako Shirouzu / Shigeyuki Yokoyama / David A Agard /   要旨: The Hsp90 molecular chaperone and its Cdc37 cochaperone help stabilize and activate more than half of the human kinome. However, both the mechanism by which these chaperones assist their "client" ...The Hsp90 molecular chaperone and its Cdc37 cochaperone help stabilize and activate more than half of the human kinome. However, both the mechanism by which these chaperones assist their "client" kinases and the reason why some kinases are addicted to Hsp90 while closely related family members are independent are unknown. Our structural understanding of these interactions is lacking, as no full-length structures of human Hsp90, Cdc37, or either of these proteins with a kinase have been elucidated. Here we report a 3.9 angstrom cryo-electron microscopy structure of the Hsp90-Cdc37-Cdk4 kinase complex. Surprisingly, the two lobes of Cdk4 are completely separated with the β4-β5 sheet unfolded. Cdc37 mimics part of the kinase N lobe, stabilizing an open kinase conformation by wedging itself between the two lobes. Finally, Hsp90 clamps around the unfolded kinase β5 strand and interacts with exposed N- and C-lobe interfaces, protecting the kinase in a trapped unfolded state. On the basis of this structure and an extensive amount of previously collected data, we propose unifying conceptual and mechanistic models of chaperone-kinase interactions. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_3341.map.gz emd_3341.map.gz | 60 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-3341-v30.xml emd-3341-v30.xml emd-3341.xml emd-3341.xml | 16.9 KB 16.9 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_3341_fsc.xml emd_3341_fsc.xml | 10.7 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_3341.tif emd_3341.tif | 135.7 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3341 http://ftp.pdbj.org/pub/emdb/structures/EMD-3341 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3341 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3341 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_3341_validation.pdf.gz emd_3341_validation.pdf.gz | 270.5 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_3341_full_validation.pdf.gz emd_3341_full_validation.pdf.gz | 269.6 KB | 表示 | |
| XML形式データ |  emd_3341_validation.xml.gz emd_3341_validation.xml.gz | 10.5 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3341 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3341 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3341 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3341 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  5fwlMC  5jwlM  3337C  3338C  3339C  3340C  3342C  3343C  3344C  5fwkC  5fwmC  5fwpC M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_3341.map.gz / 形式: CCP4 / 大きさ: 62.5 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_3341.map.gz / 形式: CCP4 / 大きさ: 62.5 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Reconstruction of Hsp90:Cdc37:Cdk4 complex. Part of series of maps, the highest resolution map being EMD-3337. This is a different subclass from the same particles as in EMD-3337, having well defined Cdk4 N-Lobe. Other relevant maps are EMD-3338, EMD-3339, EMD-3340. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.315 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
- 試料の構成要素
試料の構成要素
-全体 : Complex of Human Hsp90 beta, human Cdc37 and human Cdk4
| 全体 | 名称: Complex of Human Hsp90 beta, human Cdc37 and human Cdk4 |
|---|---|
| 要素 |
|
-超分子 #1000: Complex of Human Hsp90 beta, human Cdc37 and human Cdk4
| 超分子 | 名称: Complex of Human Hsp90 beta, human Cdc37 and human Cdk4 タイプ: sample / ID: 1000 / 詳細: All three proteins were co-expressed in Sf9 cells. 集合状態: One Hsp90 homodimer binds to one Cdc37 and one Cdk4 Number unique components: 3 |
|---|---|
| 分子量 | 実験値: 245 KDa / 理論値: 245 KDa / 手法: As cloned, verified by SDS-PAGE |
-分子 #1: Heat Shock Protein HSP 90 beta
| 分子 | 名称: Heat Shock Protein HSP 90 beta / タイプ: protein_or_peptide / ID: 1 / Name.synonym: Hsp90 / コピー数: 2 / 集合状態: Dimer / 組換発現: Yes |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) / 別称: Human / 細胞中の位置: cytoplasm Homo sapiens (ヒト) / 別称: Human / 細胞中の位置: cytoplasm |
| 分子量 | 理論値: 83 KDa |
| 組換発現 | 生物種:  組換プラスミド: pFastBacHT |
| 配列 | UniProtKB: Heat shock protein HSP 90-beta / GO: citrulline metabolic process / InterPro: Heat shock protein Hsp90 family |
-分子 #2: Hsp90 co-chaperone Cdc37
| 分子 | 名称: Hsp90 co-chaperone Cdc37 / タイプ: protein_or_peptide / ID: 2 / Name.synonym: Cdc37 / コピー数: 1 / 組換発現: Yes |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) / 別称: Human / 細胞中の位置: throughout Homo sapiens (ヒト) / 別称: Human / 細胞中の位置: throughout |
| 分子量 | 理論値: 44.5 KDa |
| 組換発現 | 生物種:  組換プラスミド: pFastBacHT |
| 配列 | UniProtKB: Hsp90 co-chaperone Cdc37 / GO: mitochondrial genome maintenance |
-分子 #3: Cyclin-dependent kinase 4
| 分子 | 名称: Cyclin-dependent kinase 4 / タイプ: protein_or_peptide / ID: 3 / Name.synonym: Cdk4 / コピー数: 1 / 組換発現: Yes |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) / 別称: Human / 細胞中の位置: throughout Homo sapiens (ヒト) / 別称: Human / 細胞中の位置: throughout |
| 分子量 | 理論値: 33.7 KDa |
| 組換発現 | 生物種:  組換プラスミド: pFastBacHT |
| 配列 | UniProtKB: Cyclin-dependent kinase 4 / GO: very long-chain fatty acid metabolic process / InterPro: Protein kinase domain |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 0.27 mg/mL |
|---|---|
| 緩衝液 | pH: 7.5 詳細: 20mM Tris-HCl (pH 7.5), 150 mM NaCl, 10 mM KCl, 10 mM MgCl2, 20 mM Na2MoO4, 2mM DTT, 0.085mM DDM |
| グリッド | 詳細: Glow discharged for 30 sec, C-flat 400 mesh 1.2/1.3 thick carbon grids (Protochips) |
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 90 % / チャンバー内温度: 95 K / 装置: FEI VITROBOT MARK III / 手法: Single blot from 4 to 6 seconds, at 20C |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| アライメント法 | Legacy - 非点収差: At high mag via FT. |
| 日付 | 2014年11月25日 |
| 撮影 | カテゴリ: CCD フィルム・検出器のモデル: GATAN K2 SUMMIT (4k x 4k) 実像数: 3718 / 平均電子線量: 44 e/Å2 / 詳細: 38 frames, 7.6 seconds total exposure / ビット/ピクセル: 8 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: OTHER / 撮影モード: BRIGHT FIELD / Cs: 2.7 mm / 最大 デフォーカス(公称値): 3.8 µm / 最小 デフォーカス(公称値): 1.4 µm / 倍率(公称値): 22500 |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
- 画像解析
画像解析
-原子モデル構築 1
| 初期モデル | PDB ID: Chain - #0 - Chain ID: A / Chain - #1 - Chain ID: B / Chain - #2 - Chain ID: E / Chain - #3 - Chain ID: K |
|---|---|
| ソフトウェア | 名称: Chimera, Rosetta |
| 詳細 | Residues 5-85 (N-lobe) of Cdk4 from 3G33 were fit in Chimera as rigid body into EMD-3341. Such fit N-lobe of Cdk4 was further tweaked in Coot in the context of 5FWK to join the Cdk4 chain and minimize clashes for each of the maps. To relieve atomic clashes or bond length/angle distortions at the linker regions, these models were subjected to "Cartesian space relax" protocol within corresponding density maps using Rosetta. Final models were selected using the combined score of Rosetta all-atom physically-realistic score and electron density score. |
| 精密化 | 空間: REAL / プロトコル: FLEXIBLE FIT / 温度因子: 200 当てはまり具合の基準: cross correlation of fit into the density with Rosetta force field score |
| 得られたモデル |  PDB-5fwl:  PDB-5jwl: |
-原子モデル構築 2
| 初期モデル | PDB ID: |
|---|---|
| ソフトウェア | 名称: Chimera, Rosetta |
| 精密化 | 空間: REAL / プロトコル: FLEXIBLE FIT / 温度因子: 200 当てはまり具合の基準: cross correlation of fit into the density with Rosetta force field score |
| 得られたモデル |  PDB-5fwl:  PDB-5jwl: |
 ムービー
ムービー コントローラー
コントローラー



































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)