[English] 日本語
 Yorodumi
Yorodumi- EMDB-31595: Cryo-EM structure of the hedgehog release protein Disp from water... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31595 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the hedgehog release protein Disp from water bear (Hypsibius dujardini) | |||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | cryo-EM / Dispatched / Hedgehog release / Hedgehog signaling / membrane protein / setrol sensing domain / water bear | |||||||||||||||||||||
| Function / homology | : / Membrane transport protein MMPL domain / MMPL family / Sterol-sensing domain (SSD) profile. / Sterol-sensing domain / smoothened signaling pathway / transmembrane transporter activity / membrane / Protein dispatched-like protein 1 Function and homology information Function and homology information | |||||||||||||||||||||
| Biological species |  Hypsibius dujardini (invertebrata) Hypsibius dujardini (invertebrata) | |||||||||||||||||||||
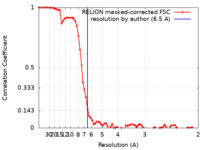
| Method | single particle reconstruction / cryo EM / Resolution: 6.5 Å | |||||||||||||||||||||
 Authors Authors | Luo Y / Wan G | |||||||||||||||||||||
| Funding support |  China, 6 items China, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Front Mol Biosci / Year: 2021 Journal: Front Mol Biosci / Year: 2021Title: Architecture of Dispatched, a Transmembrane Protein Responsible for Hedgehog Release. Authors: Yitian Luo / Guoyue Wan / Xuan Zhou / Qiuwen Wang / Yunbin Zhang / Juan Bao / Yao Cong / Yun Zhao / Dianfan Li /  Abstract: The evolutionarily conserved Hedgehog (Hh) signaling pathway is crucial for programmed cell differentiation and proliferation. Dispatched (Disp) is a 12-transmembrane protein that plays a critical ...The evolutionarily conserved Hedgehog (Hh) signaling pathway is crucial for programmed cell differentiation and proliferation. Dispatched (Disp) is a 12-transmembrane protein that plays a critical role in the Hedgehog (Hh) signaling pathway by releasing the dually lipidated ligand HhN from the membrane, a prerequisite step to the downstream signaling cascade. In this study, we focus on the Disp from water bear, a primitive animal known as the most indestructible on Earth. Using a zebrafish model, we show that the water bear homolog possesses the function of Disp. We have solved its structure to a 6.5-Å resolution using single-particle cryogenic electron microscopy. Consistent with the evolutional conservation of the pathway, the water bear Disp structure is overall similar to the previously reported structures of the fruit fly and human homologs. Although not revealing much detail at this resolution, the water bear Disp shows a different conformation compared to published structures, suggesting that they represent different functional snapshots. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31595.map.gz emd_31595.map.gz | 3.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31595-v30.xml emd-31595-v30.xml emd-31595.xml emd-31595.xml | 19.7 KB 19.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_31595_fsc.xml emd_31595_fsc.xml | 7.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_31595.png emd_31595.png | 61.4 KB | ||
| Filedesc metadata |  emd-31595.cif.gz emd-31595.cif.gz | 6.5 KB | ||
| Others |  emd_31595_half_map_1.map.gz emd_31595_half_map_1.map.gz emd_31595_half_map_2.map.gz emd_31595_half_map_2.map.gz | 26.4 MB 26.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31595 http://ftp.pdbj.org/pub/emdb/structures/EMD-31595 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31595 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31595 | HTTPS FTP |
-Related structure data
| Related structure data |  7fifMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31595.map.gz / Format: CCP4 / Size: 34.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31595.map.gz / Format: CCP4 / Size: 34.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #1
| File | emd_31595_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_31595_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Dispatched
| Entire | Name: Dispatched |
|---|---|
| Components |
|
-Supramolecule #1: Dispatched
| Supramolecule | Name: Dispatched / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Hypsibius dujardini (invertebrata) Hypsibius dujardini (invertebrata) |
| Molecular weight | Theoretical: 130 kDa/nm |
-Macromolecule #1: Protein dispatched-like protein 1
| Macromolecule | Name: Protein dispatched-like protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Hypsibius dujardini (invertebrata) Hypsibius dujardini (invertebrata) |
| Molecular weight | Theoretical: 124.943414 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGSHHHHHHH HHHHHGSPRQ LVRRRKKTVM QRYAHFLAHH PFLILTLTFC ANIVLLLVIV CTDSLPAFDD PQAGFEPRGS LINQRAQAW RNFLERDGVV QPSTRRKATT TKPVAARQNA PVQEIGTGLR NSTVKENVFS SSGMASLGPI TQPRGFFCSR P LLGHAKLI ...String: MGSHHHHHHH HHHHHGSPRQ LVRRRKKTVM QRYAHFLAHH PFLILTLTFC ANIVLLLVIV CTDSLPAFDD PQAGFEPRGS LINQRAQAW RNFLERDGVV QPSTRRKATT TKPVAARQNA PVQEIGTGLR NSTVKENVFS SSGMASLGPI TQPRGFFCSR P LLGHAKLI LRRKDAGNLL NLEGIRELCQ LDADIRRLDE FQSCSEESAD TGRLCRTWNL PNYVAVLAGK TSCHQLEEHD IR NVLNLTA TCLPFYRSGQ LKADCSTSED DGKTMQEDTI TCPGVPTDCI KDSSIYHLLY FISDKESRET GYSSMKHTLA VLP VWSSPA ALPLYRALQW GNFTRPDDDG PIQVIGMEMG LGDRLFNTAL LEDTLYVGVA AGVVVVVLWL YTGSLFVTLM NLMA IFFSL VVSYFLYVFV FRLTFFPFMN LLTCVIIVAI GADALVIFAR LWHLAKTEKD DGRFEKVVHA TFRHAAQAIL VSSLT ASAA LFSDIVNPII AIRCFSIFAG LTVLVHLFFA VTWMPACFVV ADKWGSSVYV CTKSPVCRLL YRCRCLHSGL RQYCDY FRI FFEKLLPCLV IRLKWMWMAS FVLLTVGACV VVFVFPGLQL ASGRTFSLWN SGHPSEQYRL LKDRFAFEEN RNLNNNE KV SLHFVWGVLA LNQAALLDPT DTGITTMDGR FNMSDPLSQI WMLKFCADLR QQTFFDNSTQ SDNACYFDSF MLWMETGA C SGHSQFPYPP ATFIRCVHTF SNSFPQAKHF GPNFNSLHQI DSFVLRLQTS QLFSHSYTAM QQLHQHVDQW FTAALRTAP PSLQGAWFTG DFAFFDLQQS LISGTALSLI VSLFVAFLVL FFTTLNVGVS LIAITVIAGI MLATTAALVL MEWQLSVFES TIIGLAIGL SVDFTLHYAV SYCCAESYEE RELKTNIVIS EMASCVTMSA VTTFLAGALM IPSDILFYRQ LGLFIITVTA I SLLYATIF LPACLAVLGP QGAFLQFHYP SCRPLCCRPD PSKLVEKSMH SSAEFDTTYT TGGTYDHHRE TKQCFHQSIN QP YQGKALR SPSAAANNHH VVVISATPEQ PPKTHPALLL DVDPDLADEE TDSVGELGSV PTSRGPSRGT SLIGTLEVLF Q UniProtKB: Protein dispatched-like protein 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: OTHER | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number grids imaged: 3 / Number real images: 5380 / Average exposure time: 7.6 sec. / Average electron dose: 60.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Calibrated defocus max: 3.0 µm / Calibrated defocus min: 0.9 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)