+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10452 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Drosophila melanogaster Dispatched | |||||||||
 Map data Map data | Drosophila melanogaster Dispatched | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RND transporter / transmembrane domain / ectodomain / cholesteryl hemisuccinate / detergent micelle / digitonin / monomer / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationFormation and transport of the N-HH ligand / cytoneme / wing disc pattern formation / patched ligand maturation / segment polarity determination / germ cell migration / smoothened signaling pathway / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
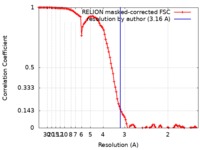
| Method | single particle reconstruction / cryo EM / Resolution: 3.16 Å | |||||||||
 Authors Authors | Korkhov VM / Cannac F | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2020 Journal: Sci Adv / Year: 2020Title: Cryo-EM structure of the Hedgehog release protein Dispatched. Authors: Fabien Cannac / Chao Qi / Julia Falschlunger / George Hausmann / Konrad Basler / Volodymyr M Korkhov /  Abstract: The Hedgehog (Hh) signaling pathway controls embryonic development and adult tissue homeostasis in multicellular organisms. In , the pathway is primed by secretion of a dually lipid-modified ...The Hedgehog (Hh) signaling pathway controls embryonic development and adult tissue homeostasis in multicellular organisms. In , the pathway is primed by secretion of a dually lipid-modified morphogen, Hh, a process dependent on a membrane-integral protein Dispatched. Although Dispatched is a critical component of the pathway, the structural basis of its activity has, so far, not been described. Here, we describe a cryo-electron microscopy structure of the Dispatched at 3.2-Å resolution. The ectodomains of Dispatched adopt an open conformation suggestive of a receptor-chaperone role. A three-dimensional reconstruction of Dispatched bound to Hh confirms the ability of Dispatched to bind Hh but using a unique mode distinct from those previously observed in structures of Hh complexes. The structure may represent the state of the complex that precedes shedding of Hh from the surface of the morphogen-releasing cell. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10452.map.gz emd_10452.map.gz | 7.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10452-v30.xml emd-10452-v30.xml emd-10452.xml emd-10452.xml | 20.3 KB 20.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10452_fsc.xml emd_10452_fsc.xml | 10.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_10452.png emd_10452.png | 55.9 KB | ||
| Filedesc metadata |  emd-10452.cif.gz emd-10452.cif.gz | 7.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10452 http://ftp.pdbj.org/pub/emdb/structures/EMD-10452 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10452 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10452 | HTTPS FTP |
-Related structure data
| Related structure data |  6tbuMC  6td6C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10452.map.gz / Format: CCP4 / Size: 107.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10452.map.gz / Format: CCP4 / Size: 107.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Drosophila melanogaster Dispatched | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.81 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Drosophila melanogaster protein Dispatched (disp)
| Entire | Name: Drosophila melanogaster protein Dispatched (disp) |
|---|---|
| Components |
|
-Supramolecule #1: Drosophila melanogaster protein Dispatched (disp)
| Supramolecule | Name: Drosophila melanogaster protein Dispatched (disp) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 139 KDa |
-Macromolecule #1: Protein dispatched
| Macromolecule | Name: Protein dispatched / type: protein_or_peptide / ID: 1 Details: Drosophila melanogaster Dispatched, tagged with a C-terminal 3C-protease cleavage site, YFP and twin-strep tag. Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 139.149875 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MLCFDSERMN WYYHVLARRP YLVVVSIAVY CVACIIVALV LNKLPDFSDP TLGFETRGTK IGERLTAWYN LLQETDHHGA LFSNPSDLW ERRRVEQGYV ETKLHPNHRR RKNKHKNRNK NKRRKEQNQS SHEHHDVAQK MMQFKKRLKA TSSPSPNLGF D TWIGDSGV ...String: MLCFDSERMN WYYHVLARRP YLVVVSIAVY CVACIIVALV LNKLPDFSDP TLGFETRGTK IGERLTAWYN LLQETDHHGA LFSNPSDLW ERRRVEQGYV ETKLHPNHRR RKNKHKNRNK NKRRKEQNQS SHEHHDVAQK MMQFKKRLKA TSSPSPNLGF D TWIGDSGV FRDYEITNDS ASSSLEPTRR TEQIEYGHNT TSVDEEEHQQ RVQTKKSTWR LLKQAATLPT DGWADMHRRQ PI EGFFCDS SPRKEYSHFV VQRIGPNATD SLFDLNGLLA MCQLQDQITE VPSYRAFCEP EMLTTECCRP WSLPNYAAML ANK SSCFDL TTEDVTSLHT LLLGCYEYFH DLKMDNHCNE IPHCRAPEEC KRLNIVFNVL NFLTDFSFIK SNDSNVYLKY AMIF IPVAQ SNRLLPLFHE WEDVELINEL VEVVAMDLGL ENELFNELLL TDVWLVSLGG TFVMASVWLY TGSAFITLMS CVAIC FSLG LAYFFYAIVL EFEFFPYMNL LAVVVIIGIG ADDVFLFLKI WHCVLTERFS NRCTLTTQSQ SALPTLENSD HTESLE NIM ALTMRHAAAS MFVTSLTTAG AFYASYSSSI TAIKCFGIFA GTVVVTNYLL MITWLPASVS IMERLFATRM SCHHPMS IK LIHACKKSIN RFCQMFEECI TKSIMNYAYL WLLIFGALGA SSAVIVFWYP GLQLPEKSHF QLFVSKHPFE VYSSLKQQ F WFEKPLQAYY NFKMHMHFVW GVQAVDDGDY TNPNSYGHLH YDNNFNVSSR PAQLWILDFC QSVRQQPFYK ETLGMLLPN CFIENLIDYM KRRCIDDMDS TRKDRSPCCD AQFPFEPHIF EYCLPQSISN MYDTTFFRPG VAGPKFAEAP RLETEDYLGM SGNESAEYS TNGSFTPLLV KALVIEFESN VAYSTIYANI RQFYESVEHW FQMQLKTAPP ELQGGWFTSD LKFYNVQDTL S HDTFVAIC LAMAASLAVL LCFTVNILIS IYAVLTVSLS IFNTVAVLIL LGWQLNILES IAVSTAIGLA VDFSLHYGIH YR MSPVKER LAATQFVLSR IIGPTVMAAT TTGLAGGIMM ASNILPYIQI GVFLVVVMIV SWFYATFFLM SLLRVAGPQH GFL ELKWPL WSKRSSGSSK FYERKPSQVI ASEQLLTPTS SAIVELANSE THELESLNSN SLIKTISGIE SAHALSSLPR DFEH SFQTM HECKYQTYPS TSN UniProtKB: Protein dispatched |
-Macromolecule #3: CHOLESTEROL HEMISUCCINATE
| Macromolecule | Name: CHOLESTEROL HEMISUCCINATE / type: ligand / ID: 3 / Number of copies: 9 / Formula: Y01 |
|---|---|
| Molecular weight | Theoretical: 486.726 Da |
| Chemical component information |  ChemComp-Y01: |
-Macromolecule #4: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 4 / Number of copies: 1 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Number real images: 9216 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)