+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31585 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of AtTPC1 with 1 mM Ca2+ | ||||||||||||
 Map data Map data | Structure of AtTPC1 with 1 mM Ca2 | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | non-selective cation channel / dimer / vacuole / TRANSPORT PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of jasmonic acid biosynthetic process / seed germination / regulation of stomatal movement / plant-type vacuole / vacuole / vacuolar membrane / monoatomic ion channel complex / voltage-gated calcium channel activity / bioluminescence / generation of precursor metabolites and energy ...regulation of jasmonic acid biosynthetic process / seed germination / regulation of stomatal movement / plant-type vacuole / vacuole / vacuolar membrane / monoatomic ion channel complex / voltage-gated calcium channel activity / bioluminescence / generation of precursor metabolites and energy / calcium-mediated signaling / calcium ion transport / calcium ion binding / Golgi apparatus / identical protein binding / plasma membrane / cytosol Similarity search - Function | ||||||||||||
| Biological species |   Human respiratory syncytial virus Human respiratory syncytial virus | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | ||||||||||||
 Authors Authors | Ye F / Xu L | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2021 Journal: Proc Natl Acad Sci U S A / Year: 2021Title: Voltage-gating and cytosolic Ca activation mechanisms of two-pore channel AtTPC1. Authors: Fan Ye / Lingyi Xu / Xiaoxiao Li / Weizhong Zeng / Ninghai Gan / Cheng Zhao / Wei Yang / Youxing Jiang / Jiangtao Guo /   Abstract: two-pore channel AtTPC1 is a voltage-gated, Ca-modulated, nonselective cation channel that is localized in the vacuolar membrane and responsible for generating slow vacuolar (SV) current. Under ... two-pore channel AtTPC1 is a voltage-gated, Ca-modulated, nonselective cation channel that is localized in the vacuolar membrane and responsible for generating slow vacuolar (SV) current. Under depolarizing membrane potential, cytosolic Ca activates AtTPC1 by binding at the EF-hand domain, whereas luminal Ca inhibits the channel by stabilizing the voltage-sensing domain II (VSDII) in the resting state. Here, we present 2.8 to 3.3 Å cryoelectron microscopy (cryo-EM) structures of AtTPC1 in two conformations, one in closed conformation with unbound EF-hand domain and resting VSDII and the other in a partially open conformation with Ca-bound EF-hand domain and activated VSDII. Structural comparison between the two different conformations allows us to elucidate the structural mechanisms of voltage gating, cytosolic Ca activation, and their coupling in AtTPC1. This study also provides structural insight into the general voltage-gating mechanism among voltage-gated ion channels. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31585.map.gz emd_31585.map.gz | 31.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31585-v30.xml emd-31585-v30.xml emd-31585.xml emd-31585.xml | 11.8 KB 11.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_31585.png emd_31585.png | 166.6 KB | ||
| Filedesc metadata |  emd-31585.cif.gz emd-31585.cif.gz | 5.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31585 http://ftp.pdbj.org/pub/emdb/structures/EMD-31585 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31585 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31585 | HTTPS FTP |
-Validation report
| Summary document |  emd_31585_validation.pdf.gz emd_31585_validation.pdf.gz | 484.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_31585_full_validation.pdf.gz emd_31585_full_validation.pdf.gz | 483.9 KB | Display | |
| Data in XML |  emd_31585_validation.xml.gz emd_31585_validation.xml.gz | 6.1 KB | Display | |
| Data in CIF |  emd_31585_validation.cif.gz emd_31585_validation.cif.gz | 6.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31585 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31585 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31585 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-31585 | HTTPS FTP |
-Related structure data
| Related structure data |  7fhkMC  7fhlC  7fhnC  7fhoC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31585.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31585.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of AtTPC1 with 1 mM Ca2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.014 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
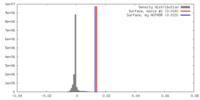
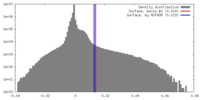
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : AtTPC1 homodimer
| Entire | Name: AtTPC1 homodimer |
|---|---|
| Components |
|
-Supramolecule #1: AtTPC1 homodimer
| Supramolecule | Name: AtTPC1 homodimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Two pore calcium channel protein 1,GFP
| Macromolecule | Name: Two pore calcium channel protein 1,GFP / type: protein_or_peptide / ID: 1 Details: The fusion protein of AtTPC1 (UNP residues 1-733), LINKER and GFP (UNP residues 2-239) Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human respiratory syncytial virus Human respiratory syncytial virus |
| Molecular weight | Theoretical: 114.644297 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MEDPLIGRDS LGGGGTDRVR RSEAITHGTP FQKAAALVDL AEDGIGLPVE ILDQSSFGES ARYYFIFTRL DLIWSLNYFA LLFLNFFEQ PLWCEKNPKP SCKDRDYYYL GELPYLTNAE SIIYEVITLA ILLVHTFFPI SYEGSRIFWT SRLNLVKVAC V VILFVDVL ...String: MEDPLIGRDS LGGGGTDRVR RSEAITHGTP FQKAAALVDL AEDGIGLPVE ILDQSSFGES ARYYFIFTRL DLIWSLNYFA LLFLNFFEQ PLWCEKNPKP SCKDRDYYYL GELPYLTNAE SIIYEVITLA ILLVHTFFPI SYEGSRIFWT SRLNLVKVAC V VILFVDVL VDFLYLSPLA FDFLPFRIAP YVRVIIFILS IRELRDTLVL LSGMLGTYLN ILALWMLFLL FASWIAFVMF ED TQQGLTV FTSYGATLYQ MFILFTTSNN PDVWIPAYKS SRWSSVFFVL YVLIGVYFVT NLILAVVYDS FKEQLAKQVS GMD QMKRRM LEKAFGLIDS DKNGEIDKNQ CIKLFEQLTN YRTLPKISKE EFGLIFDELD DTRDFKINKD EFADLCQAIA LRFQ KEEVP SLFEHFPQIY HSALSQQLRA FVRSPNFGYA ISFILIINFI AVVVETTLDI EESSAQKPWQ VAEFVFGWIY VLEMA LKIY TYGFENYWRE GANRFDFLVT WVIVIGETAT FITPDENTFF SNGEWIRYLL LARMLRLIRL LMNVQRYRAF IATFIT LIP SLMPYLGTIF CVLCIYCSIG VQVFGGLVNA GNKKLFETEL AEDDYLLFNF NDYPNGMVTL FNLLVMGNWQ VWMESYK DL TGTWWSITYF VSFYVITILL LLNLVVAFVL EAFFTELDLE EEEKCQGQDS QEKRNRRRSA GSKSRSQRVD TLLHHMLG D ELSKPECSTS DTSTAGLVPR GSAAAAVSKG EELFTGVVPI LVELDGDVNG HKFSVSGEGE GDATYGKLTL KFICTTGKL PVPWPTLVTT LTYGVQCFSR YPDHMKQHDF FKSAMPEGYV QERTIFFKDD GNYKTRAEVK FEGDTLVNRI ELKGIDFKED GNILGHKLE YNYNSHNVYI MADKQKNGIK VNFKIRHNIE DGSVQLADHY QQNTPIGDGP VLLPDNHYLS TQSKLSKDPN E KRDHMVLL EFVTAAGITL GMDELYKSGL RSHHHHHHHH UniProtKB: Two pore calcium channel protein 1, GFP |
-Macromolecule #2: AtTPC1-Cter
| Macromolecule | Name: AtTPC1-Cter / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1.039273 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) |
-Macromolecule #3: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 3 / Number of copies: 4 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 64.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.3 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 1106424 |
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: OTHER |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)