[English] 日本語
 Yorodumi
Yorodumi- EMDB-31258: STRUCTURE OF PHOTOSYNTHETIC LH1-RC SUPER-COMPLEX OF RHODOSPIRILLU... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-31258 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | STRUCTURE OF PHOTOSYNTHETIC LH1-RC SUPER-COMPLEX OF RHODOSPIRILLUM RUBRUM | |||||||||||||||||||||
 Map data Map data | LH1-RC complex of Rsp. rubrum | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
 Keywords Keywords | LH1-RC COMPLEX / PHOTOSYNTHESIS / PURPLE BACTERIA | |||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationorganelle inner membrane / plasma membrane-derived chromatophore membrane / plasma membrane light-harvesting complex / : / bacteriochlorophyll binding / photosynthetic electron transport in photosystem II / photosynthesis, light reaction / metal ion binding / plasma membrane Similarity search - Function | |||||||||||||||||||||
| Biological species |  Rhodospirillum rubrum (bacteria) / Rhodospirillum rubrum (bacteria) /  Rhodospirillum rubrum (strain ATCC 11170 / ATH 1.1.1 / DSM 467 / LMG 4362 / NCIB 8255 / S1) (bacteria) Rhodospirillum rubrum (strain ATCC 11170 / ATH 1.1.1 / DSM 467 / LMG 4362 / NCIB 8255 / S1) (bacteria) | |||||||||||||||||||||
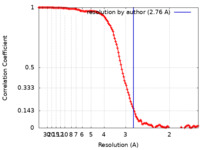
| Method | single particle reconstruction / cryo EM / Resolution: 2.76 Å | |||||||||||||||||||||
 Authors Authors | Tani K / Kanno R | |||||||||||||||||||||
| Funding support |  Japan, 6 items Japan, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Biochemistry / Year: 2021 Journal: Biochemistry / Year: 2021Title: Cryo-EM Structure of the Photosynthetic LH1-RC Complex from . Authors: Kazutoshi Tani / Ryo Kanno / Xuan-Cheng Ji / Malgorzata Hall / Long-Jiang Yu / Yukihiro Kimura / Michael T Madigan / Akira Mizoguchi / Bruno M Humbel / Zheng-Yu Wang-Otomo /    Abstract: () is one of the most widely used model organisms in bacterial photosynthesis. This purple phototroph is characterized by the presence of both rhodoquinone (RQ) and ubiquinone as electron carriers ... () is one of the most widely used model organisms in bacterial photosynthesis. This purple phototroph is characterized by the presence of both rhodoquinone (RQ) and ubiquinone as electron carriers and bacteriochlorophyll (BChl) esterified at the propionic acid side chain by geranylgeraniol (BChl ) instead of phytol. Despite intensive efforts, the structure of the light-harvesting-reaction center (LH1-RC) core complex from remains at low resolutions. Using cryo-EM, here we present a robust new view of the LH1-RC at 2.76 Å resolution. The LH1 complex forms a closed, slightly elliptical ring structure with 16 αβ-polypeptides surrounding the RC. Our biochemical analysis detected RQ molecules in the purified LH1-RC, and the cryo-EM density map specifically positions RQ at the Q site in the RC. The geranylgeraniol side chains of BChl coordinated by LH1 β-polypeptides exhibit a highly homologous tail-up conformation that allows for interactions with the bacteriochlorin rings of nearby LH1 α-associated BChls . The structure also revealed key protein-protein interactions in both N- and C-terminal regions of the LH1 αβ-polypeptides, mainly within a face-to-face structural subunit. Our high-resolution LH1-RC structure provides new insight for evaluating past experimental and computational results obtained with this old organism over many decades and lays the foundation for more detailed exploration of light-energy conversion, quinone transport, and structure-function relationships in this pigment-protein complex. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_31258.map.gz emd_31258.map.gz | 166.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-31258-v30.xml emd-31258-v30.xml emd-31258.xml emd-31258.xml | 21.9 KB 21.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_31258_fsc.xml emd_31258_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_31258.png emd_31258.png | 168.8 KB | ||
| Filedesc metadata |  emd-31258.cif.gz emd-31258.cif.gz | 7.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-31258 http://ftp.pdbj.org/pub/emdb/structures/EMD-31258 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31258 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-31258 | HTTPS FTP |
-Related structure data
| Related structure data |  7eqdMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_31258.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_31258.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | LH1-RC complex of Rsp. rubrum | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : Photosynthetic LH1-RC complex from the purple phototrophic bacter...
+Supramolecule #1: Photosynthetic LH1-RC complex from the purple phototrophic bacter...
+Macromolecule #1: Reaction center protein L chain
+Macromolecule #2: Reaction center protein M chain
+Macromolecule #3: Photoreaction center protein H
+Macromolecule #4: Light-harvesting protein B-870 alpha chain
+Macromolecule #5: Light-harvesting protein B-870 beta chain
+Macromolecule #6: Trans-Geranyl BACTERIOCHLOROPHYLL A
+Macromolecule #7: Trans-Geranyl BACTERIOPHEOPHYTIN A
+Macromolecule #8: UBIQUINONE-10
+Macromolecule #9: (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-...
+Macromolecule #10: DODECYL-BETA-D-MALTOSIDE
+Macromolecule #11: FE (III) ION
+Macromolecule #12: 2-azanyl-5-[(2~{E},6~{E},8~{E},10~{E},12~{E},14~{E},18~{E},22~{E}...
+Macromolecule #13: SPIRILLOXANTHIN
+Macromolecule #14: CARDIOLIPIN
+Macromolecule #15: DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.0 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Model: Quantifoil R2/2 / Material: MOLYBDENUM / Mesh: 200 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Chamber temperature: 277 K / Instrument: LEICA EM GP |
| Details | This sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average exposure time: 30.6 sec. / Average electron dose: 42.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Overall B value: 60 / Target criteria: Correlation coefficient |
| Output model |  PDB-7eqd: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)