[English] 日本語
 Yorodumi
Yorodumi- EMDB-30636: Cryo-EM structure of human XKR8-basigin complex bound to Fab fragment -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30636 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human XKR8-basigin complex bound to Fab fragment | ||||||||||||||||||||||||
 Map data Map data | Cryo-EM map for human XKR8/Basigin complex bound to Fab fragment | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | XKR8 / basigin / scramblase / phospholipid / TRANSPORT PROTEIN | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationDefective SLC16A1 causes symptomatic deficiency in lactate transport (SDLT) / Proton-coupled monocarboxylate transport / phosphatidylserine exposure on apoptotic cell surface / tolerance induction to self antigen / positive regulation of matrix metallopeptidase secretion / dendrite self-avoidance / acrosomal membrane / neutrophil clearance / phospholipid scramblase activity / cell-cell adhesion mediator activity ...Defective SLC16A1 causes symptomatic deficiency in lactate transport (SDLT) / Proton-coupled monocarboxylate transport / phosphatidylserine exposure on apoptotic cell surface / tolerance induction to self antigen / positive regulation of matrix metallopeptidase secretion / dendrite self-avoidance / acrosomal membrane / neutrophil clearance / phospholipid scramblase activity / cell-cell adhesion mediator activity / endothelial tube morphogenesis / response to mercury ion / engulfment of apoptotic cell / neural retina development / apoptotic process involved in development / photoreceptor cell maintenance / Basigin interactions / Aspirin ADME / odontogenesis of dentin-containing tooth / homophilic cell-cell adhesion / D-mannose binding / decidualization / positive regulation of vascular endothelial growth factor production / photoreceptor outer segment / response to cAMP / Integrin cell surface interactions / positive regulation of myoblast differentiation / Degradation of the extracellular matrix / neutrophil chemotaxis / photoreceptor inner segment / positive regulation of endothelial cell migration / embryo implantation / axon guidance / protein localization to plasma membrane / establishment of localization in cell / sarcolemma / response to peptide hormone / positive regulation of interleukin-6 production / melanosome / signaling receptor activity / virus receptor activity / angiogenesis / basolateral plasma membrane / positive regulation of viral entry into host cell / cell surface receptor signaling pathway / endosome / cadherin binding / Golgi membrane / axon / focal adhesion / endoplasmic reticulum membrane / perinuclear region of cytoplasm / mitochondrion / extracellular exosome / membrane / plasma membrane Similarity search - Function | ||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | ||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | ||||||||||||||||||||||||
 Authors Authors | Sakuragi T / Kanai R | ||||||||||||||||||||||||
| Funding support |  Japan, 7 items Japan, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2021 Journal: Nat Struct Mol Biol / Year: 2021Title: The tertiary structure of the human Xkr8-Basigin complex that scrambles phospholipids at plasma membranes. Authors: Takaharu Sakuragi / Ryuta Kanai / Akihisa Tsutsumi / Hirotaka Narita / Eriko Onishi / Kohei Nishino / Takuya Miyazaki / Takeshi Baba / Hidetaka Kosako / Atsushi Nakagawa / Masahide Kikkawa / ...Authors: Takaharu Sakuragi / Ryuta Kanai / Akihisa Tsutsumi / Hirotaka Narita / Eriko Onishi / Kohei Nishino / Takuya Miyazaki / Takeshi Baba / Hidetaka Kosako / Atsushi Nakagawa / Masahide Kikkawa / Chikashi Toyoshima / Shigekazu Nagata /  Abstract: Xkr8-Basigin is a plasma membrane phospholipid scramblase activated by kinases or caspases. We combined cryo-EM and X-ray crystallography to investigate its structure at an overall resolution of 3. ...Xkr8-Basigin is a plasma membrane phospholipid scramblase activated by kinases or caspases. We combined cryo-EM and X-ray crystallography to investigate its structure at an overall resolution of 3.8 Å. Its membrane-spanning region carrying 22 charged amino acids adopts a cuboid-like structure stabilized by salt bridges between hydrophilic residues in transmembrane helices. Phosphatidylcholine binding was observed in a hydrophobic cleft on the surface exposed to the outer leaflet of the plasma membrane. Six charged residues placed from top to bottom inside the molecule were essential for scrambling phospholipids in inward and outward directions, apparently providing a pathway for their translocation. A tryptophan residue was present between the head group of phosphatidylcholine and the extracellular end of the path. Its mutation to alanine made the Xkr8-Basigin complex constitutively active, indicating that it plays a vital role in regulating its scramblase activity. The structure of Xkr8-Basigin provides insights into the molecular mechanisms underlying phospholipid scrambling. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30636.map.gz emd_30636.map.gz | 10.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30636-v30.xml emd-30636-v30.xml emd-30636.xml emd-30636.xml | 16.3 KB 16.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_30636.png emd_30636.png | 57.5 KB | ||
| Filedesc metadata |  emd-30636.cif.gz emd-30636.cif.gz | 6.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30636 http://ftp.pdbj.org/pub/emdb/structures/EMD-30636 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30636 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30636 | HTTPS FTP |
-Related structure data
| Related structure data |  7dceMC  7d9zC  7daaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-11046 (Title: Human Xkr8-Basigin complex bound to Fab fragment / Data size: 451.5 / Data #1: Unaligned K3 movies [micrographs - multiframe]) EMPIAR-11046 (Title: Human Xkr8-Basigin complex bound to Fab fragment / Data size: 451.5 / Data #1: Unaligned K3 movies [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30636.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30636.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map for human XKR8/Basigin complex bound to Fab fragment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.245 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
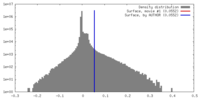
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : human XKR8-basigin complex bound to Fab fragment
| Entire | Name: human XKR8-basigin complex bound to Fab fragment |
|---|---|
| Components |
|
-Supramolecule #1: human XKR8-basigin complex bound to Fab fragment
| Supramolecule | Name: human XKR8-basigin complex bound to Fab fragment / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Molecular weight | Theoretical: 110 KDa |
-Supramolecule #2: human XKR8-basigin complex
| Supramolecule | Name: human XKR8-basigin complex / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: Fab fragment
| Supramolecule | Name: Fab fragment / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3-#4 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Isoform 2 of Basigin
| Macromolecule | Name: Isoform 2 of Basigin / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 19.592814 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AGPPRVKAVK SSEHINEGET AMLVCKSESV PPVTDWAWYK ITDSEDKALM QGSESRFFVS SSQGRSELHI ENLNMEADPG QYRCQGTSS KGSDQAIITL RVRSHLAALW PFLGIVAEVL VLVTIIFIYE KRRKPEDVLD DDDAGSAPLK SSGQHQNDKG K NVRQRNSS DYKDDDDK UniProtKB: Basigin |
-Macromolecule #2: XK-related protein 8
| Macromolecule | Name: XK-related protein 8 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 45.975609 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPWSSRGALL RDLVLGVLGT AAFLLDLGTD LWAAVQYALG GRYLWAALVL ALLGLASVAL QLFSWLWLRA DPAGLHGSQP PRRCLALLH LLQLGYLYRC VQELRQGLLV WQQEEPSEFD LAYADFLALD ISMLRLFETF LETAPQLTLV LAIMLQSGRA E YYQWVGIC ...String: MPWSSRGALL RDLVLGVLGT AAFLLDLGTD LWAAVQYALG GRYLWAALVL ALLGLASVAL QLFSWLWLRA DPAGLHGSQP PRRCLALLH LLQLGYLYRC VQELRQGLLV WQQEEPSEFD LAYADFLALD ISMLRLFETF LETAPQLTLV LAIMLQSGRA E YYQWVGIC TSFLGISWAL LDYHRALRTC LPSKPLLGLG SSVIYFLWNL LLLWPRVLAV ALFSALFPSY VALHFLGLWL VL LLWVWLQ GTDFMPDPSS EWLYRVTVAT ILYFSWFNVA EGRTRGRAII HFAFLLSDSI LLVATWVTHS SWLPSGIPLQ LWL PVGCGC FFLGLALRLV YYHWLHPSCC WKPDPDQVDG ARSLLSPEGY QLPQNRRMTH LAQKFFPKAK DEAASPVKGV DEFE NLYFQ UniProtKB: XK-related protein 8 |
-Macromolecule #3: Heavy chain of Fab fragment
| Macromolecule | Name: Heavy chain of Fab fragment / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 22.869639 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: (PCA)SVEESGGRL VTPGTPLTLT CTVSGFSLSD YAMNWVRQAP GKGLEWIGII YASGSRYYAS WAKGRFTISK TSTTVD LKI TSPTTEDTAT YFCARYYAGS DIWGPGTLVT VSSASTKGPS VFPLAPSSKS TSGGTAALGC LVKDYFPEPV TVSWNSG AL TSGVHTFPAV ...String: (PCA)SVEESGGRL VTPGTPLTLT CTVSGFSLSD YAMNWVRQAP GKGLEWIGII YASGSRYYAS WAKGRFTISK TSTTVD LKI TSPTTEDTAT YFCARYYAGS DIWGPGTLVT VSSASTKGPS VFPLAPSSKS TSGGTAALGC LVKDYFPEPV TVSWNSG AL TSGVHTFPAV LQSSGLYSLS SVVTVPSSSL GTQTYICNVN HKPSNTKVDK KVEPKSCDK |
-Macromolecule #4: Light chain of Fab fragment
| Macromolecule | Name: Light chain of Fab fragment / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.261865 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ADVVMTQTPS SVSAAVGGTV TINCQASQSI SAYLAWYQQK PGQPPKLLIY DASDLASGVS SRFKGSGSGT QFTLTISALE CADAATYYC QSYYAIITYG AAFGGGTEVV VKRTVAAPSV FIFPPSDEQL KSGTASVVCL LNNFYPREAK VQWKVDNALQ S GNSQESVT ...String: ADVVMTQTPS SVSAAVGGTV TINCQASQSI SAYLAWYQQK PGQPPKLLIY DASDLASGVS SRFKGSGSGT QFTLTISALE CADAATYYC QSYYAIITYG AAFGGGTEVV VKRTVAAPSV FIFPPSDEQL KSGTASVVCL LNNFYPREAK VQWKVDNALQ S GNSQESVT EQDSKDCTYS LSSTLTLSKA DYEKHKVYAC EVTHQGLSSP VTKSFNRGEC |
-Macromolecule #5: 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE
| Macromolecule | Name: 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE / type: ligand / ID: 5 / Number of copies: 1 / Formula: DLP |
|---|---|
| Molecular weight | Theoretical: 782.082 Da |
| Chemical component information |  ChemComp-DLP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.8 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 62124 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)