[English] 日本語
 Yorodumi
Yorodumi- EMDB-22281: CryoEM structure of human presequence protease in partial closed ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22281 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of human presequence protease in partial closed state 1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Partial open state / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationMitochondrial protein import / amyloid-beta complex / growth cone lamellipodium / cellular response to norepinephrine stimulus / growth cone filopodium / microglia development / collateral sprouting in absence of injury / Hydrolases; Acting on peptide bonds (peptidases); Metalloendopeptidases / Formyl peptide receptors bind formyl peptides and many other ligands / axo-dendritic transport ...Mitochondrial protein import / amyloid-beta complex / growth cone lamellipodium / cellular response to norepinephrine stimulus / growth cone filopodium / microglia development / collateral sprouting in absence of injury / Hydrolases; Acting on peptide bonds (peptidases); Metalloendopeptidases / Formyl peptide receptors bind formyl peptides and many other ligands / axo-dendritic transport / regulation of Wnt signaling pathway / axon midline choice point recognition / astrocyte activation involved in immune response / regulation of synapse structure or activity / NMDA selective glutamate receptor signaling pathway / regulation of spontaneous synaptic transmission / mating behavior / growth factor receptor binding / peptidase activator activity / Golgi-associated vesicle / : / PTB domain binding / positive regulation of amyloid fibril formation / Insertion of tail-anchored proteins into the endoplasmic reticulum membrane / astrocyte projection / Lysosome Vesicle Biogenesis / neuron remodeling / Deregulated CDK5 triggers multiple neurodegenerative pathways in Alzheimer's disease models / nuclear envelope lumen / dendrite development / positive regulation of protein metabolic process / TRAF6 mediated NF-kB activation / signaling receptor activator activity / negative regulation of long-term synaptic potentiation / Advanced glycosylation endproduct receptor signaling / modulation of excitatory postsynaptic potential / main axon / transition metal ion binding / The NLRP3 inflammasome / regulation of multicellular organism growth / intracellular copper ion homeostasis / ECM proteoglycans / regulation of presynapse assembly / positive regulation of T cell migration / neuronal dense core vesicle / Purinergic signaling in leishmaniasis infection / positive regulation of chemokine production / Notch signaling pathway / cellular response to manganese ion / clathrin-coated pit / extracellular matrix organization / neuron projection maintenance / astrocyte activation / ionotropic glutamate receptor signaling pathway / Mitochondrial protein degradation / positive regulation of calcium-mediated signaling / positive regulation of mitotic cell cycle / axonogenesis / protein serine/threonine kinase binding / response to interleukin-1 / platelet alpha granule lumen / cellular response to copper ion / cellular response to cAMP / positive regulation of glycolytic process / central nervous system development / dendritic shaft / trans-Golgi network membrane / endosome lumen / positive regulation of long-term synaptic potentiation / positive regulation of interleukin-1 beta production / adult locomotory behavior / learning / positive regulation of JNK cascade / Post-translational protein phosphorylation / locomotory behavior / serine-type endopeptidase inhibitor activity / enzyme activator activity / microglial cell activation / positive regulation of non-canonical NF-kappaB signal transduction / cellular response to nerve growth factor stimulus / TAK1-dependent IKK and NF-kappa-B activation / regulation of long-term neuronal synaptic plasticity / metalloendopeptidase activity / synapse organization / recycling endosome / protein processing / visual learning / response to lead ion / positive regulation of interleukin-6 production / Golgi lumen / cognition / Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) / endocytosis / cellular response to amyloid-beta / neuron projection development / metallopeptidase activity / positive regulation of inflammatory response / positive regulation of tumor necrosis factor production / Platelet degranulation / heparin binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
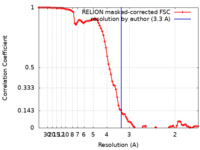
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Liang WG / Zhao M | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: Structural basis for the mechanisms of human presequence protease conformational switch and substrate recognition. Authors: Wenguang G Liang / Juwina Wijaya / Hui Wei / Alex J Noble / Jordan M Mancl / Swansea Mo / David Lee / John V Lin King / Man Pan / Chang Liu / Carla M Koehler / Minglei Zhao / Clinton S ...Authors: Wenguang G Liang / Juwina Wijaya / Hui Wei / Alex J Noble / Jordan M Mancl / Swansea Mo / David Lee / John V Lin King / Man Pan / Chang Liu / Carla M Koehler / Minglei Zhao / Clinton S Potter / Bridget Carragher / Sheng Li / Wei-Jen Tang /  Abstract: Presequence protease (PreP), a 117 kDa mitochondrial M16C metalloprotease vital for mitochondrial proteostasis, degrades presequence peptides cleaved off from nuclear-encoded proteins and other ...Presequence protease (PreP), a 117 kDa mitochondrial M16C metalloprotease vital for mitochondrial proteostasis, degrades presequence peptides cleaved off from nuclear-encoded proteins and other aggregation-prone peptides, such as amyloid β (Aβ). PreP structures have only been determined in a closed conformation; thus, the mechanisms of substrate binding and selectivity remain elusive. Here, we leverage advanced vitrification techniques to overcome the preferential denaturation of one of two ~55 kDa homologous domains of PreP caused by air-water interface adsorption. Thereby, we elucidate cryoEM structures of three apo-PreP open states along with Aβ- and citrate synthase presequence-bound PreP at 3.3-4.6 Å resolution. Together with integrative biophysical and pharmacological approaches, these structures reveal the key stages of the PreP catalytic cycle and how the binding of substrates or PreP inhibitor drives a rigid body motion of the protein for substrate binding and catalysis. Together, our studies provide key mechanistic insights into M16C metalloproteases for future therapeutic innovations. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22281.map.gz emd_22281.map.gz | 49.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22281-v30.xml emd-22281-v30.xml emd-22281.xml emd-22281.xml | 24.6 KB 24.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22281_fsc.xml emd_22281_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_22281.png emd_22281.png | 164.8 KB | ||
| Masks |  emd_22281_msk_1.map emd_22281_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-22281.cif.gz emd-22281.cif.gz | 7.1 KB | ||
| Others |  emd_22281_additional_1.map.gz emd_22281_additional_1.map.gz emd_22281_half_map_1.map.gz emd_22281_half_map_1.map.gz emd_22281_half_map_2.map.gz emd_22281_half_map_2.map.gz | 59.9 MB 49.6 MB 49.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22281 http://ftp.pdbj.org/pub/emdb/structures/EMD-22281 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22281 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22281 | HTTPS FTP |
-Related structure data
| Related structure data |  6xovMC  6xosC  6xotC  6xouC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10937 (Title: CryoEM of PreP prepared via Chameleon / Data size: 2.2 TB / Data #1: Apo-PreP micrographs [micrographs - multiframe] / Data #2: PreP half particles [micrographs - single frame] EMPIAR-10937 (Title: CryoEM of PreP prepared via Chameleon / Data size: 2.2 TB / Data #1: Apo-PreP micrographs [micrographs - multiframe] / Data #2: PreP half particles [micrographs - single frame]Data #3: Citrate synthase presequence bound PreP [micrographs - multiframe] Data #4: Amyloid beta bound PreP [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22281.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22281.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.855 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_22281_msk_1.map emd_22281_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_22281_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_22281_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_22281_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : CryoEM map of Human Presequence Protease in partial close state 1
| Entire | Name: CryoEM map of Human Presequence Protease in partial close state 1 |
|---|---|
| Components |
|
-Supramolecule #1: CryoEM map of Human Presequence Protease in partial close state 1
| Supramolecule | Name: CryoEM map of Human Presequence Protease in partial close state 1 type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Presequence protease, mitochondrial
| Macromolecule | Name: Presequence protease, mitochondrial / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on peptide bonds (peptidases); Metalloendopeptidases |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 114.901461 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHAAA CERALQYKLG DKIHGFTVNQ VTSVPELFLT AVKLTHDDTG ARYLHLARED TNNLFSVQFR TTPMDSTGVP HILEHTVLC GSQKYPCRDP FFKMLNRSLS TFMNAFTASD YTLYPFSTQN PKDFQNLLSV YLDATFFPCL RELDFWQEGW R LEHENPSD ...String: MHHHHHHAAA CERALQYKLG DKIHGFTVNQ VTSVPELFLT AVKLTHDDTG ARYLHLARED TNNLFSVQFR TTPMDSTGVP HILEHTVLC GSQKYPCRDP FFKMLNRSLS TFMNAFTASD YTLYPFSTQN PKDFQNLLSV YLDATFFPCL RELDFWQEGW R LEHENPSD PQTPLVFKGV VFNEMKGAFT DNERIFSQHL QNRLLPDHTY SVVSGGDPLC IPELTWEQLK QFHATHYHPS NA RFFTYGN FPLEQHLKQI HEEALSKFQK IEPSTVVPAQ TPWDKPREFQ ITCGPDSFAT DPSKQTTVSV SFLLPDITDT FEA FTLSLL SSLLTSGPNS PFYKALIESG LGTDFSPDVG YNGYTREAYF SVGLQGIVEK DIETVRSLID RTIDEVVEKG FEDD RIEAL LHKIEIQMKH QSTSFGLMLT SYIASCWNHD GDPVELLKLG NQLAKFRQCL QENPKFLQEK VKQYFKNNQH KLTLS MRPD DKYHEKQAQV EATKLKQKVE ALSPGDRQQI YEKGLELRSQ QSKPQDASCL PALKVSDIEP TIPVTELDVV LTAGDI PVQ YCAQPTNGMV YFRAFSSLNT LPEELRPYVP LFCSVLTKLG CGLLDYREQA QQIELKTGGM SASPHVLPDD SHMDTYE QG VLFSSLCLDR NLPDMMQLWS EIFNNPCFEE EEHFKVLVKM TAQELANGIP DSGHLYASIR AGRTLTPAGD LQETFSGM D QVRLMKRIAE MTDIKPILRK LPRIKKHLLN GDNMRCSVNA TPQQMPQTEK AVEDFLRSIG RSKKERRPVR PHTVEKPVP SSSGGDAHVP HGSQVIRKLV MEPTFKPWQM KTHFLMPFPV NYVGECIRTV PYTDPDHASL KILARLMTAK FLHTEIREKG GAYGGGAKL SHNGIFTLYS YRDPNTIETL QSFGKAVDWA KSGKFTQQDI DEAKLSVFST VDAPVAPSDK GMDHFLYGLS D EMKQAHRE QLFAVSHDKL LAVSDRYLGT GKSTHGLAIL GPENPKIAKD PSWIIR UniProtKB: Presequence protease, mitochondrial |
-Macromolecule #2: Amyloid-beta precursor protein
| Macromolecule | Name: Amyloid-beta precursor protein / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 4.335852 KDa |
| Sequence | String: DAEFRHDSGY EVHHQKLVFF AEDVGSNKGA IIGLMVGGVV UniProtKB: Amyloid-beta precursor protein |
-Macromolecule #3: Amyloid-beta precursor protein
| Macromolecule | Name: Amyloid-beta precursor protein / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 273.33 Da |
| Sequence | String: (UNK)(UNK)(UNK) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.7 Details: 20 mM Tris, pH 7.7, 150 mM NaCl, 10mM KCl, 20 mM EDTA and 1 mM 2-mercaptoethanol |
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 308 K / Instrument: SPOTITON |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average exposure time: 6.0 sec. / Average electron dose: 66.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)