[English] 日本語
 Yorodumi
Yorodumi- EMDB-22282: CryoEM structure of human presequence protease in partial close s... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22282 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of human presequence protease in partial close state 2, induced by presequence of citrate synthase | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Partial open state / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationMitochondrial protein import / Hydrolases; Acting on peptide bonds (peptidases); Metalloendopeptidases / : / enzyme activator activity / protein processing / metalloendopeptidase activity / metallopeptidase activity / mitochondrial matrix / mitochondrion / proteolysis / zinc ion binding Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
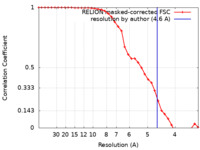
| Method | single particle reconstruction / cryo EM / Resolution: 4.6 Å | |||||||||
 Authors Authors | Liang WG / Tang W | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: CryoEM structure of human presequence protease in partial open state 2, induced by presequence of citrate synthase Authors: Liang WG / Zhao M / Tang W | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22282.map.gz emd_22282.map.gz | 2.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22282-v30.xml emd-22282-v30.xml emd-22282.xml emd-22282.xml | 20.7 KB 20.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22282_fsc.xml emd_22282_fsc.xml | 3.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_22282.png emd_22282.png | 108.9 KB | ||
| Masks |  emd_22282_msk_1.map emd_22282_msk_1.map | 3.4 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-22282.cif.gz emd-22282.cif.gz | 6.3 KB | ||
| Others |  emd_22282_additional_1.map.gz emd_22282_additional_1.map.gz emd_22282_half_map_1.map.gz emd_22282_half_map_1.map.gz emd_22282_half_map_2.map.gz emd_22282_half_map_2.map.gz | 3.2 MB 2.4 MB 2.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22282 http://ftp.pdbj.org/pub/emdb/structures/EMD-22282 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22282 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22282 | HTTPS FTP |
-Related structure data
| Related structure data |  6xowMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10937 (Title: CryoEM of PreP prepared via Chameleon / Data size: 2.2 TB / Data #1: Apo-PreP micrographs [micrographs - multiframe] / Data #2: PreP half particles [micrographs - single frame] EMPIAR-10937 (Title: CryoEM of PreP prepared via Chameleon / Data size: 2.2 TB / Data #1: Apo-PreP micrographs [micrographs - multiframe] / Data #2: PreP half particles [micrographs - single frame]Data #3: Citrate synthase presequence bound PreP [micrographs - multiframe] Data #4: Amyloid beta bound PreP [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22282.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22282.map.gz / Format: CCP4 / Size: 3.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.71 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_22282_msk_1.map emd_22282_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_22282_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_22282_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_22282_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : CryoEM map of Human Presequence Protease in partial close state 2...
| Entire | Name: CryoEM map of Human Presequence Protease in partial close state 2, induced by presequence of citrate synthase |
|---|---|
| Components |
|
-Supramolecule #1: CryoEM map of Human Presequence Protease in partial close state 2...
| Supramolecule | Name: CryoEM map of Human Presequence Protease in partial close state 2, induced by presequence of citrate synthase type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Presequence protease, mitochondrial
| Macromolecule | Name: Presequence protease, mitochondrial / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on peptide bonds (peptidases); Metalloendopeptidases |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 114.901461 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHAAA CERALQYKLG DKIHGFTVNQ VTSVPELFLT AVKLTHDDTG ARYLHLARED TNNLFSVQFR TTPMDSTGVP HILEHTVLC GSQKYPCRDP FFKMLNRSLS TFMNAFTASD YTLYPFSTQN PKDFQNLLSV YLDATFFPCL RELDFWQEGW R LEHENPSD ...String: MHHHHHHAAA CERALQYKLG DKIHGFTVNQ VTSVPELFLT AVKLTHDDTG ARYLHLARED TNNLFSVQFR TTPMDSTGVP HILEHTVLC GSQKYPCRDP FFKMLNRSLS TFMNAFTASD YTLYPFSTQN PKDFQNLLSV YLDATFFPCL RELDFWQEGW R LEHENPSD PQTPLVFKGV VFNEMKGAFT DNERIFSQHL QNRLLPDHTY SVVSGGDPLC IPELTWEQLK QFHATHYHPS NA RFFTYGN FPLEQHLKQI HEEALSKFQK IEPSTVVPAQ TPWDKPREFQ ITCGPDSFAT DPSKQTTVSV SFLLPDITDT FEA FTLSLL SSLLTSGPNS PFYKALIESG LGTDFSPDVG YNGYTREAYF SVGLQGIVEK DIETVRSLID RTIDEVVEKG FEDD RIEAL LHKIEIQMKH QSTSFGLMLT SYIASCWNHD GDPVELLKLG NQLAKFRQCL QENPKFLQEK VKQYFKNNQH KLTLS MRPD DKYHEKQAQV EATKLKQKVE ALSPGDRQQI YEKGLELRSQ QSKPQDASCL PALKVSDIEP TIPVTELDVV LTAGDI PVQ YCAQPTNGMV YFRAFSSLNT LPEELRPYVP LFCSVLTKLG CGLLDYREQA QQIELKTGGM SASPHVLPDD SHMDTYE QG VLFSSLCLDR NLPDMMQLWS EIFNNPCFEE EEHFKVLVKM TAQELANGIP DSGHLYASIR AGRTLTPAGD LQETFSGM D QVRLMKRIAE MTDIKPILRK LPRIKKHLLN GDNMRCSVNA TPQQMPQTEK AVEDFLRSIG RSKKERRPVR PHTVEKPVP SSSGGDAHVP HGSQVIRKLV MEPTFKPWQM KTHFLMPFPV NYVGECIRTV PYTDPDHASL KILARLMTAK FLHTEIREKG GAYGGGAKL SHNGIFTLYS YRDPNTIETL QSFGKAVDWA KSGKFTQQDI DEAKLSVFST VDAPVAPSDK GMDHFLYGLS D EMKQAHRE QLFAVSHDKL LAVSDRYLGT GKSTHGLAIL GPENPKIAKD PSWIIR UniProtKB: Presequence protease, mitochondrial |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.7 Details: 20 mM Tris, pH 7.7, 150 mM NaCl, 10mM KCl, 20 mM EDTA and 1 mM 2-mercaptoethanol |
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 308 K / Instrument: SPOTITON |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average exposure time: 6.0 sec. / Average electron dose: 66.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)