+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22070 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Legionella pneumophila Dot/Icm T4SS | ||||||||||||||||||||||||
 Map data Map data | Dot/Icm T4SS | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | Secretion / T4SS / Dot / PROTEIN TRANSPORT | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology information | ||||||||||||||||||||||||
| Biological species |  | ||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | ||||||||||||||||||||||||
 Authors Authors | Durie CL / Sheedlo MJ | ||||||||||||||||||||||||
| Funding support |  United States, 7 items United States, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Elife / Year: 2020 Journal: Elife / Year: 2020Title: Structural analysis of the Dot/Icm type IV secretion system core complex. Authors: Clarissa L Durie / Michael J Sheedlo / Jeong Min Chung / Brenda G Byrne / Min Su / Thomas Knight / Michele Swanson / D Borden Lacy / Melanie D Ohi /  Abstract: is an opportunistic pathogen that causes the potentially fatal pneumonia Legionnaires' Disease. This infection and subsequent pathology require the Dot/Icm Type IV Secretion System (T4SS) to deliver ... is an opportunistic pathogen that causes the potentially fatal pneumonia Legionnaires' Disease. This infection and subsequent pathology require the Dot/Icm Type IV Secretion System (T4SS) to deliver effector proteins into host cells. Compared to prototypical T4SSs, the Dot/Icm assembly is much larger, containing ~27 different components including a core complex reported to be composed of five proteins: DotC, DotD, DotF, DotG, and DotH. Using single particle cryo-electron microscopy (cryo-EM), we report reconstructions of the core complex of the Dot/Icm T4SS that includes a symmetry mismatch between distinct structural features of the outer membrane cap (OMC) and periplasmic ring (PR). We present models of known core complex proteins, DotC, DotD, and DotH, and two structurally similar proteins within the core complex, DotK and Lpg0657. This analysis reveals the stoichiometry and contact interfaces between the key proteins of the Dot/Icm T4SS core complex and provides a framework for understanding a complex molecular machine. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22070.map.gz emd_22070.map.gz | 429.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22070-v30.xml emd-22070-v30.xml emd-22070.xml emd-22070.xml | 16.5 KB 16.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_22070.png emd_22070.png | 83.4 KB | ||
| Filedesc metadata |  emd-22070.cif.gz emd-22070.cif.gz | 6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22070 http://ftp.pdbj.org/pub/emdb/structures/EMD-22070 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22070 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22070 | HTTPS FTP |
-Related structure data
| Related structure data |  6x65MC  6x62C  6x64C  6x66C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10893 (Title: Motion corrected micrographs - purified Dot/Icm T4SS particles EMPIAR-10893 (Title: Motion corrected micrographs - purified Dot/Icm T4SS particlesData size: 190.4 Data #1: Motion corrected micrographs Legionella pneumophila Dot/Icm T4SS [micrographs - single frame]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22070.map.gz / Format: CCP4 / Size: 506 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22070.map.gz / Format: CCP4 / Size: 506 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Dot/Icm T4SS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.64 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
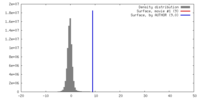
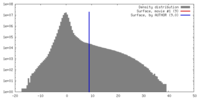
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Legionella pneumophila Dot/Icm T4SS
| Entire | Name: Legionella pneumophila Dot/Icm T4SS |
|---|---|
| Components |
|
-Supramolecule #1: Legionella pneumophila Dot/Icm T4SS
| Supramolecule | Name: Legionella pneumophila Dot/Icm T4SS / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3, #5 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Type IV secretion system unknown protein fragment
| Macromolecule | Name: Type IV secretion system unknown protein fragment / type: protein_or_peptide / ID: 1 / Number of copies: 75 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 19.421865 KDa |
| Sequence | String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) ...String: (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK) |
-Macromolecule #2: DotC
| Macromolecule | Name: DotC / type: protein_or_peptide / ID: 2 / Number of copies: 13 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 34.162441 KDa |
| Sequence | String: MRKFILSLSI LLSALLVACS SRNHYGDTGS LAGLQAMADS KYTRAQKKQK MGKIREMALK ETALSVGAQA GLAWRAKIID EQLNKQARN LDAIYDFNSL VLEHNILPPV LLEGRNTLNL ADAQSIRISD RTYKVAKQAH FITTPPTWRQ YLWMDYVKPE A PNVTLLPK ...String: MRKFILSLSI LLSALLVACS SRNHYGDTGS LAGLQAMADS KYTRAQKKQK MGKIREMALK ETALSVGAQA GLAWRAKIID EQLNKQARN LDAIYDFNSL VLEHNILPPV LLEGRNTLNL ADAQSIRISD RTYKVAKQAH FITTPPTWRQ YLWMDYVKPE A PNVTLLPK TKAEKEIWCI YTERGWKNGI DQANTILEEN IARIKEDFGG MILYRKLLAM NMVSPPYVSH TDLGVTGDGS EI HIDDRVL RITALPELNV NSAEWRAAVA KDENALERFK NMEKLANQAK IVITNKSWQP IIAPVS UniProtKB: DotC |
-Macromolecule #3: DotD
| Macromolecule | Name: DotD / type: protein_or_peptide / ID: 3 / Number of copies: 26 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 17.860678 KDa |
| Sequence | String: MNNNKIVIMF IFSALLAGCA GTMKFKKPPI NNPSDDATIK LAEAAVSVSD SMLEMAKVEK VITPPSKDNT LTIPNAYNLQ ARASVDWSG PIEELTARIA KAAHFRFRVL GKSPSVPVLI SISTKDESLA EILRDIDYQA GKKASIHVYP NSQVVELRYA K IYS UniProtKB: DotD |
-Macromolecule #4: Type IV secretion protein IcmK
| Macromolecule | Name: Type IV secretion protein IcmK / type: protein_or_peptide / ID: 4 / Number of copies: 13 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 38.958926 KDa |
| Sequence | String: MMKKYDQLCK YCLVIGLTFS MSCSIYAADQ SDDAQQALQQ LRMLQQKLSQ NPSPDAQSGA GDGGDNAASD STQQPNQSGQ ANAPAANQT ATAGGDGQII SQDDAEVIDK KAFKDMTRNL YPLNPEQVVK LKQIYETSEY AKAATPGTPP KPTATSQFVN L SPGSTPPV ...String: MMKKYDQLCK YCLVIGLTFS MSCSIYAADQ SDDAQQALQQ LRMLQQKLSQ NPSPDAQSGA GDGGDNAASD STQQPNQSGQ ANAPAANQT ATAGGDGQII SQDDAEVIDK KAFKDMTRNL YPLNPEQVVK LKQIYETSEY AKAATPGTPP KPTATSQFVN L SPGSTPPV IRLSQGFVSS LVFLDSTGAP WPIAAYDLGD PSSFNIQWDK TSNTLMIQAT KLYNYGNLAV RLRGLNTPVM LT LIPGQKA VDYRVDLRVQ GYGPNAKSMP TEEGIPPSAN DLLLHVLEGV PPPGSRRLVV SGGDARAWLS NEKMYVRTNL TIL SPGWLA SMTSADGTHA YEMQKSPVLL VSWHGKVMQL KVEGL UniProtKB: Type IV secretion protein IcmK |
-Macromolecule #5: Inner membrane lipoprotein YiaD
| Macromolecule | Name: Inner membrane lipoprotein YiaD / type: protein_or_peptide / ID: 5 / Number of copies: 13 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 21.096492 KDa |
| Sequence | String: MRSLRTNYIY VLFKTTGLLF LLLLSACNRS GYIPENEVPK LPCRVDGACD ATIIKMMTDL NKKGIKVASV GQNYLISIPA SALFADQSP RLNWASYSLL NEIAAFLKQF RKIAITVTSY SSKYVSVKRE RALTLARSRV VSEYLWSQGV DSRIIFTQGL G SDKPITSY TLGGDRSPNA RVEITFRRAV A UniProtKB: LphA (DotK) |
-Macromolecule #6: Outer membrane protein, OmpA family protein
| Macromolecule | Name: Outer membrane protein, OmpA family protein / type: protein_or_peptide / ID: 6 / Number of copies: 13 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.582047 KDa |
| Sequence | String: MRNLMRCLIM IKSLIKGVDM SRKLAKTRIL GYGLMICFLA GCFHPPYNNF QPDRRAVKRV GVDTGIGAVA GAIASGTASG TLIGAAAGG TVGLVASIYR DSKRKIIRDL QKQDIQYVEY GDTRTLIIPT DKYFMFSSPR LNEICYPGLN NVIRLLNFYP Q STIYVAGF ...String: MRNLMRCLIM IKSLIKGVDM SRKLAKTRIL GYGLMICFLA GCFHPPYNNF QPDRRAVKRV GVDTGIGAVA GAIASGTASG TLIGAAAGG TVGLVASIYR DSKRKIIRDL QKQDIQYVEY GDTRTLIIPT DKYFMFSSPR LNEICYPGLN NVIRLLNFYP Q STIYVAGF TDNVGSRSHK RKLSQAQAET MMTFLWANGI AAKRLKAEGY GDKNAISDNA IIHGSAQNRR IEIQWFTSPA QP PQPQMAY VK UniProtKB: Outer membrane protein, OmpA family protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 58.1 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)