+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Integrative Structure of the human intron lariat Spliceosome (ILS'') | |||||||||
 マップデータ マップデータ | composite map | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | mRNA / splicing / intron lariat spliceosome / pre-mRNA | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報RNA lariat debranching enzyme activator activity / : / post-spliceosomal complex / U2-type post-mRNA release spliceosomal complex / negative regulation of double-strand break repair via nonhomologous end joining / biomineral tissue development / spliceosomal complex disassembly / protection from non-homologous end joining at telomere / regulation of skeletal muscle satellite cell proliferation / positive regulation of myoblast proliferation ...RNA lariat debranching enzyme activator activity / : / post-spliceosomal complex / U2-type post-mRNA release spliceosomal complex / negative regulation of double-strand break repair via nonhomologous end joining / biomineral tissue development / spliceosomal complex disassembly / protection from non-homologous end joining at telomere / regulation of skeletal muscle satellite cell proliferation / positive regulation of myoblast proliferation / regulation of retinoic acid receptor signaling pathway / post-mRNA release spliceosomal complex / U2 snRNP binding / endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / U7 snRNA binding / histone pre-mRNA DCP binding / 3'-5' RNA helicase activity / U7 snRNP / generation of catalytic spliceosome for first transesterification step / histone methyltransferase binding / regulation of vitamin D receptor signaling pathway / histone pre-mRNA 3'end processing complex / SLBP independent Processing of Histone Pre-mRNAs / SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs / protein methylation / U12-type spliceosomal complex / embryonic brain development / methylosome / nuclear retinoic acid receptor binding / 7-methylguanosine cap hypermethylation / positive regulation of androgen receptor activity / Prp19 complex / U1 snRNP binding / pICln-Sm protein complex / mRNA 3'-end processing / ATP-dependent activity, acting on RNA / sno(s)RNA-containing ribonucleoprotein complex / snRNP binding / U2-type catalytic step 1 spliceosome / RNA splicing, via transesterification reactions / small nuclear ribonucleoprotein complex / pre-mRNA binding / SMN-Sm protein complex / response to alkaloid / telomerase RNA binding / spliceosomal tri-snRNP complex / telomerase holoenzyme complex / U2-type spliceosomal complex / U2-type precatalytic spliceosome / positive regulation by host of viral transcription / mRNA cis splicing, via spliceosome / P granule / positive regulation of vitamin D receptor signaling pathway / commitment complex / U2-type prespliceosome assembly / nuclear vitamin D receptor binding / Transport of Mature mRNA derived from an Intron-Containing Transcript / U2-type catalytic step 2 spliceosome / Notch binding / Regulation of gene expression in late stage (branching morphogenesis) pancreatic bud precursor cells / U4 snRNP / positive regulation of mRNA splicing, via spliceosome / RUNX3 regulates NOTCH signaling / U2 snRNP / NOTCH4 Intracellular Domain Regulates Transcription / RNA Polymerase II Transcription Termination / U1 snRNP / NOTCH3 Intracellular Domain Regulates Transcription / ubiquitin-ubiquitin ligase activity / positive regulation of neurogenesis / WD40-repeat domain binding / U2-type prespliceosome / lipid biosynthetic process / nuclear androgen receptor binding / snoRNA binding / precatalytic spliceosome / K63-linked polyubiquitin modification-dependent protein binding / muscle organ development / cyclosporin A binding / Notch-HLH transcription pathway / generation of catalytic spliceosome for second transesterification step / positive regulation of transforming growth factor beta receptor signaling pathway / Formation of paraxial mesoderm / mRNA Splicing - Minor Pathway / spliceosomal complex assembly / SMAD binding / mitotic G2 DNA damage checkpoint signaling / protein K63-linked ubiquitination / mRNA 3'-splice site recognition / protein peptidyl-prolyl isomerization / blastocyst development / protein localization to nucleus / spliceosomal tri-snRNP complex assembly / intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator / U5 snRNA binding / U5 snRNP / transcription-coupled nucleotide-excision repair / negative regulation of protein-containing complex assembly / positive regulation of G1/S transition of mitotic cell cycle / embryonic organ development 類似検索 - 分子機能 | |||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.5 Å | |||||||||
 データ登録者 データ登録者 | Rothe P / Vorlaender MK / Plaschka C | |||||||||
| 資金援助 | European Union, 1件
| |||||||||
 引用 引用 |  ジャーナル: Nature / 年: 2024 ジャーナル: Nature / 年: 2024タイトル: Mechanism for the initiation of spliceosome disassembly. 著者: Matthias K Vorländer / Patricia Rothe / Justus Kleifeld / Eric D Cormack / Lalitha Veleti / Daria Riabov-Bassat / Laura Fin / Alex W Phillips / Luisa Cochella / Clemens Plaschka /   要旨: Precursor-mRNA (pre-mRNA) splicing requires the assembly, remodelling and disassembly of the multi-megadalton ribonucleoprotein complex called the spliceosome. Recent studies have shed light on ...Precursor-mRNA (pre-mRNA) splicing requires the assembly, remodelling and disassembly of the multi-megadalton ribonucleoprotein complex called the spliceosome. Recent studies have shed light on spliceosome assembly and remodelling for catalysis, but the mechanism of disassembly remains unclear. Here we report cryo-electron microscopy structures of nematode and human terminal intron lariat spliceosomes along with biochemical and genetic data. Our results uncover how four disassembly factors and the conserved RNA helicase DHX15 initiate spliceosome disassembly. The disassembly factors probe large inner and outer spliceosome surfaces to detect the release of ligated mRNA. Two of these factors, TFIP11 and C19L1, and three general spliceosome subunits, SYF1, SYF2 and SDE2, then dock and activate DHX15 on the catalytic U6 snRNA to initiate disassembly. U6 therefore controls both the start and end of pre-mRNA splicing. Taken together, our results explain the molecular basis of the initiation of canonical spliceosome disassembly and provide a framework to understand general spliceosomal RNA helicase control and the discard of aberrant spliceosomes. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_19399.map.gz emd_19399.map.gz | 8 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-19399-v30.xml emd-19399-v30.xml emd-19399.xml emd-19399.xml | 61.3 KB 61.3 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_19399.png emd_19399.png | 96.2 KB | ||
| Filedesc metadata |  emd-19399.cif.gz emd-19399.cif.gz | 18.7 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-19399 http://ftp.pdbj.org/pub/emdb/structures/EMD-19399 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19399 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-19399 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_19399_validation.pdf.gz emd_19399_validation.pdf.gz | 374.7 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_19399_full_validation.pdf.gz emd_19399_full_validation.pdf.gz | 374.3 KB | 表示 | |
| XML形式データ |  emd_19399_validation.xml.gz emd_19399_validation.xml.gz | 7.1 KB | 表示 | |
| CIF形式データ |  emd_19399_validation.cif.gz emd_19399_validation.cif.gz | 8.2 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19399 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19399 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19399 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-19399 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  8ro2MC  8ro0C  8ro1C  9fmdC  50477  50478  50479  50480  50481  50482  50483  50484  50485  50486  50487  50488  50489  50490 M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_19399.map.gz / 形式: CCP4 / 大きさ: 282.6 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_19399.map.gz / 形式: CCP4 / 大きさ: 282.6 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | composite map | ||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.2375 Å | ||||||||||||||||||||||||||||||||||||
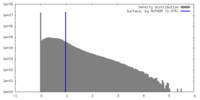
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
- 試料の構成要素
試料の構成要素
+全体 : Human Intron-lariat splicoesome
+超分子 #1: Human Intron-lariat splicoesome
+分子 #1: U2 snRNA
+分子 #2: U6 snRNA
+分子 #26: U5 snRNA
+分子 #29: INTRON
+分子 #3: 116 kDa U5 small nuclear ribonucleoprotein component
+分子 #4: Pre-mRNA-splicing factor ISY1 homolog
+分子 #5: ATP-dependent RNA helicase DHX15
+分子 #6: U5 small nuclear ribonucleoprotein 40 kDa protein
+分子 #7: Crooked neck-like protein 1
+分子 #8: Pre-mRNA-splicing factor SPF27
+分子 #9: CWF19-like protein 1
+分子 #10: Protein BUD31 homolog
+分子 #11: Pre-mRNA-splicing factor RBM22
+分子 #12: Intron-binding protein aquarius
+分子 #13: SNW domain-containing protein 1
+分子 #14: Peptidyl-prolyl cis-trans isomerase-like 1
+分子 #15: Pre-mRNA-processing factor 17
+分子 #16: Coiled-coil domain-containing protein 12
+分子 #17: Small nuclear ribonucleoprotein Sm D3
+分子 #18: Small nuclear ribonucleoprotein Sm D1
+分子 #19: Small nuclear ribonucleoprotein Sm D2
+分子 #20: Small nuclear ribonucleoprotein E
+分子 #21: Small nuclear ribonucleoprotein F
+分子 #22: Small nuclear ribonucleoprotein G
+分子 #23: Pre-mRNA-processing factor 19
+分子 #24: Splicing regulator SDE2
+分子 #25: Splicing factor ESS-2 homolog
+分子 #27: Pre-mRNA-processing-splicing factor 8
+分子 #28: Pre-mRNA-splicing factor SYF1
+分子 #30: Cell division cycle 5-like protein
+分子 #31: CWF19-like protein 2
+分子 #32: Pre-mRNA-splicing factor SYF2
+分子 #33: Spliceosome-associated protein CWC15 homolog
+分子 #34: PAX3- and PAX7-binding protein 1
+分子 #35: Pleiotropic regulator 1
+分子 #36: Tuftelin-interacting protein 11
+分子 #37: Small nuclear ribonucleoprotein-associated proteins B and B'
+分子 #38: GUANOSINE-5'-TRIPHOSPHATE
+分子 #39: ZINC ION
+分子 #40: INOSITOL HEXAKISPHOSPHATE
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.9 |
|---|---|
| 凍結 | 凍結剤: ETHANE |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: FEI FALCON IV (4k x 4k) 平均電子線量: 50.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): 2.0 µm / 最小 デフォーカス(公称値): 0.75 µm |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
- 画像解析
画像解析
| 初期モデル | モデルのタイプ: PDB ENTRY PDBモデル - PDB ID: |
|---|---|
| 最終 再構成 | 解像度のタイプ: BY AUTHOR / 解像度: 3.5 Å / 解像度の算出法: FSC 0.143 CUT-OFF / 使用した粒子像数: 87951 |
| 初期 角度割当 | タイプ: OTHER |
| 最終 角度割当 | タイプ: OTHER |
 ムービー
ムービー コントローラー
コントローラー





















































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)