+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12521 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | MycP5-free ESX-5 inner membrane complex, state I | |||||||||||||||
 Map data Map data | -50 Bfactor sharpening of 3D refinement map. | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | T7SS / mycobacteria / protein transport / secretion / type VII secretion system / membrane / MEMBRANE PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationHydrolases; Acting on acid anhydrides / peptidoglycan-based cell wall / hydrolase activity / ATP hydrolysis activity / DNA binding / ATP binding / plasma membrane Similarity search - Function | |||||||||||||||
| Biological species |  Mycobacterium tuberculosis H37Rv (bacteria) / Mycobacterium tuberculosis H37Rv (bacteria) /  Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) | |||||||||||||||
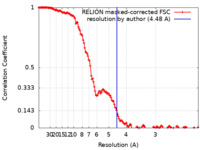
| Method | single particle reconstruction / cryo EM / Resolution: 4.48 Å | |||||||||||||||
 Authors Authors | Bunduc CM / Fahrenkamp D / Wald J / Ummels R / Bitter W / Houben ENG / Marlovits TC | |||||||||||||||
| Funding support |  Germany, Germany,  Netherlands, European Union, 4 items Netherlands, European Union, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: Structure and dynamics of a mycobacterial type VII secretion system. Authors: Catalin M Bunduc / Dirk Fahrenkamp / Jiri Wald / Roy Ummels / Wilbert Bitter / Edith N G Houben / Thomas C Marlovits /   Abstract: Mycobacterium tuberculosis is the cause of one of the most important infectious diseases in humans, which leads to 1.4 million deaths every year. Specialized protein transport systems-known as ...Mycobacterium tuberculosis is the cause of one of the most important infectious diseases in humans, which leads to 1.4 million deaths every year. Specialized protein transport systems-known as type VII secretion systems (T7SSs)-are central to the virulence of this pathogen, and are also crucial for nutrient and metabolite transport across the mycobacterial cell envelope. Here we present the structure of an intact T7SS inner-membrane complex of M. tuberculosis. We show how the 2.32-MDa ESX-5 assembly, which contains 165 transmembrane helices, is restructured and stabilized as a trimer of dimers by the MycP protease. A trimer of MycP caps a central periplasmic dome-like chamber that is formed by three EccB dimers, with the proteolytic sites of MycP facing towards the cavity. This chamber suggests a central secretion and processing conduit. Complexes without MycP show disruption of the EccB periplasmic assembly and increased flexibility, which highlights the importance of MycP for complex integrity. Beneath the EccB-MycP chamber, dimers of the EccC ATPase assemble into three bundles of four transmembrane helices each, which together seal the potential central secretion channel. Individual cytoplasmic EccC domains adopt two distinctive conformations that probably reflect different secretion states. Our work suggests a previously undescribed mechanism of protein transport and provides a structural scaffold to aid in the development of drugs against this major human pathogen. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12521.map.gz emd_12521.map.gz | 19.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12521-v30.xml emd-12521-v30.xml emd-12521.xml emd-12521.xml | 24.3 KB 24.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12521_fsc.xml emd_12521_fsc.xml | 14.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_12521.png emd_12521.png | 77.9 KB | ||
| Masks |  emd_12521_msk_1.map emd_12521_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-12521.cif.gz emd-12521.cif.gz | 7.2 KB | ||
| Others |  emd_12521_additional_1.map.gz emd_12521_additional_1.map.gz emd_12521_additional_2.map.gz emd_12521_additional_2.map.gz emd_12521_half_map_1.map.gz emd_12521_half_map_1.map.gz emd_12521_half_map_2.map.gz emd_12521_half_map_2.map.gz | 193.3 MB 39.9 MB 194.4 MB 194.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12521 http://ftp.pdbj.org/pub/emdb/structures/EMD-12521 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12521 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12521 | HTTPS FTP |
-Validation report
| Summary document |  emd_12521_validation.pdf.gz emd_12521_validation.pdf.gz | 883.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_12521_full_validation.pdf.gz emd_12521_full_validation.pdf.gz | 883 KB | Display | |
| Data in XML |  emd_12521_validation.xml.gz emd_12521_validation.xml.gz | 21.1 KB | Display | |
| Data in CIF |  emd_12521_validation.cif.gz emd_12521_validation.cif.gz | 27.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12521 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12521 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12521 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-12521 | HTTPS FTP |
-Related structure data
| Related structure data |  7npuMC  7np7C  7nprC  7npsC  7nptC  7npvC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_12521.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12521.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | -50 Bfactor sharpening of 3D refinement map. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
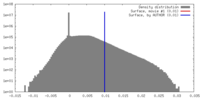
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_12521_msk_1.map emd_12521_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
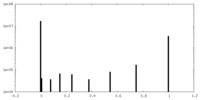
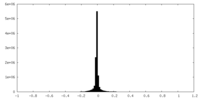
| Density Histograms |
-Additional map: 3D refinement map.
| File | emd_12521_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D refinement map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
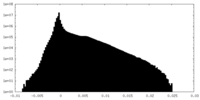
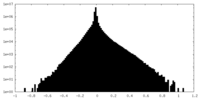
| Density Histograms |
-Additional map: #1
| File | emd_12521_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half2 of 3D refinement map.
| File | emd_12521_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half2 of 3D refinement map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half1 of 3D refinement map.
| File | emd_12521_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half1 of 3D refinement map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : MycP5-free ESX-5 inner membrane complex, state I
| Entire | Name: MycP5-free ESX-5 inner membrane complex, state I |
|---|---|
| Components |
|
-Supramolecule #1: MycP5-free ESX-5 inner membrane complex, state I
| Supramolecule | Name: MycP5-free ESX-5 inner membrane complex, state I / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis H37Rv (bacteria) Mycobacterium tuberculosis H37Rv (bacteria) |
-Macromolecule #1: ESX-5 secretion system ATPase EccB5
| Macromolecule | Name: ESX-5 secretion system ATPase EccB5 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: Hydrolases; Acting on acid anhydrides |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria)Strain: ATCC 25618 / H37Rv |
| Molecular weight | Theoretical: 53.769988 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Sequence | String: MAEESRGQRG SGYGLGLSTR TQVTGYQFLA RRTAMALTRW RVRMEIEPGR RQTLAVVASV SAALVICLGA LLWSFISPSG QLNESPIIA DRDSGALYVR VGDRLYPALN LASARLITGR PDNPHLVRSS QIATMPRGPL VGIPGAPSSF SPKSPPASSW L VCDTVATS ...String: MAEESRGQRG SGYGLGLSTR TQVTGYQFLA RRTAMALTRW RVRMEIEPGR RQTLAVVASV SAALVICLGA LLWSFISPSG QLNESPIIA DRDSGALYVR VGDRLYPALN LASARLITGR PDNPHLVRSS QIATMPRGPL VGIPGAPSSF SPKSPPASSW L VCDTVATS SSIGSLQGVT VTVIDGTPDL TGHRQILSGS DAVVLRYGGD AWVIREGRRS RIEPTNRAVL LPLGLTPEQV SQ ARPMSRA LFDALPVGPE LLVPEVPNAG GPATFPGAPG PIGTVIVTPQ ISGPQQYSLV LGDGVQTLPP LVAQILQNAG SAG NTKPLT VEPSTLAKMP VVNRLDLSAY PDNPLEVVDI REHPSTCWWW ERTAGENRAR VRVVSGPTIP VAATEMNKVV SLVK ADTSG RQADQVYFGP DHANFVAVTG NNPGAQTSES LWWVTDAGAR FGVEDSKEAR DALGLTLTPS LAPWVALRLL PQGPT LSRA DALVEHDTLP MDMTPAELVV PK UniProtKB: ESX-5 secretion system ATPase EccB5 |
-Macromolecule #2: ESX-5 secretion system protein EccC5
| Macromolecule | Name: ESX-5 secretion system protein EccC5 / type: protein_or_peptide / ID: 2 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria)Strain: ATCC 25618 / H37Rv |
| Molecular weight | Theoretical: 152.90075 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Sequence | String: MKRGFARPTP EKPPVIKPEN IVLSTPLSIP PPEGKPWWLI VVGVVVVGLL GGMVAMVFAS GSHVFGGIGS IFPLFMMVGI MMMMFRGMG GGQQQMSRPK LDAMRAQFML MLDMLRETAQ ESADSMDANY RWFHPAPNTL AAAVGSPRMW ERKPDGKDLN F GVVRVGVG ...String: MKRGFARPTP EKPPVIKPEN IVLSTPLSIP PPEGKPWWLI VVGVVVVGLL GGMVAMVFAS GSHVFGGIGS IFPLFMMVGI MMMMFRGMG GGQQQMSRPK LDAMRAQFML MLDMLRETAQ ESADSMDANY RWFHPAPNTL AAAVGSPRMW ERKPDGKDLN F GVVRVGVG MTRPEVTWGE PQNMPTDIEL EPVTGKALQE FGRYQSVVYN LPKMVSLLVE PWYALVGERE QVLGLMRAII CQ LAFSHGP DHVQMIVVSS DLDQWDWVKW LPHFGDSRRH DAAGNARMVY TSVREFAAEQ AELFAGRGSF TPRHASSSAQ TPT PHTVII ADVDDPQWEY VISAEGVDGV TFFDLTGSSM WTDIPERKLQ FDKTGVIEAL PRDRDTWMVI DDKAWFFALT DQVS IAEAE EFAQKLAQWR LAEAYEEIGQ RVAHIGARDI LSYYGIDDPG NIDFDSLWAS RTDTMGRSRL RAPFGNRSDN GELLF LDMK SLDEGGDGPH GVMSGTTGSG KSTLVRTVIE SLMLSHPPEE LQFVLADLKG GSAVKPFAGV PHVSRIITDL EEDQAL MER FLDALWGEIA RRKAICDSAG VDDAKEYNSV RARMRARGQD MAPLPMLVVV IDEFYEWFRI MPTAVDVLDS IGRQGRA YW IHLMMASQTI ESRAEKLMEN MGYRLVLKAR TAGAAQAAGV PNAVNLPAQA GLGYFRKSLE DIIRFQAEFL WRDYFQPG V SIDGEEAPAL VHSIDYIRPQ LFTNSFTPLE VSVGGPDIEP VVAQPNGEVL ESDDIEGGED EDEEGVRTPK VGTVIIDQL RKIKFEPYRL WQPPLTQPVA IDDLVNRFLG RPWHKEYGSA CNLVFPIGII DRPYKHDQPP WTVDTSGPGA NVLILGAGGS GKTTALQTL ICSAALTHTP QQVQFYCLAY SSTALTTVSR IPHVGEVAGP TDPYGVRRTV AELLALVRER KRSFLECGIA S MEMFRRRK FGGEAGPVPD DGFGDVYLVI DNYRALAEEN EVLIEQVNVI INQGPSFGVH VVVTADRESE LRPPVRSGFG SR IELRLAA VEDAKLVRSR FAKDVPVKPG RGMVAVNYVR LDSDPQAGLH TLVARPALGS TPDNVFECDS VVAAVSRLTS AQA PPVRRL PARFGVEQVR ELASRDTRQG VGAGGIAWAI SELDLAPVYL NFAENSHLMV TGRRECGRTT TLATIMSEIG RLYA PGASS APPPAPGRPS AQVWLVDPRR QLLTALGSDY VERFAYNLDG VVAMMGELAA ALAGREPPPG LSAEELLSRS WWSGP EIFL IVDDIQQLPP GFDSPLHKAV PFVNRAADVG LHVIVTRTFG GWSSAGSDPM LRALHQANAP LLVMDADPDE GFIRGK MKG GPLPRGRGLL MAEDTGVFVQ VAATEVRR UniProtKB: ESX-5 secretion system protein EccC5 |
-Macromolecule #3: ESX-5 secretion system protein EccD5
| Macromolecule | Name: ESX-5 secretion system protein EccD5 / type: protein_or_peptide / ID: 3 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria)Strain: ATCC 25618 / H37Rv |
| Molecular weight | Theoretical: 53.480906 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Sequence | String: MTAVADAPQA DIEGVASPQA VVVGVMAGEG VQIGVLLDAN APVSVMTDPL LKVVNSRLRE LGEAPLEATG RGRWALCLVD GAPLRATQS LTEQDVYDGD RLWIRFIADT ERRSQVIEHI STAVASDLSK RFARIDPIVA VQVGASMVAT GVVLATGVLG W WRWHHNTW ...String: MTAVADAPQA DIEGVASPQA VVVGVMAGEG VQIGVLLDAN APVSVMTDPL LKVVNSRLRE LGEAPLEATG RGRWALCLVD GAPLRATQS LTEQDVYDGD RLWIRFIADT ERRSQVIEHI STAVASDLSK RFARIDPIVA VQVGASMVAT GVVLATGVLG W WRWHHNTW LTTIYTAVIG VLVLAVAMLL LMRAKTDADR RVADIMLMSA IMPVTVAAAA APPGPVGSPQ AVLGFGVLTV AA ALALRFT GRRLGIYTTI VIIGALTMLA ALARMVAATS AVTLLSSLLL ICVVAYHAAP ALSRRLAGIR LPVFPSATSR WVF EARPDL PTTVVVSGGS APVLEGPSSV RDVLLQAERA RSFLSGLLTG LGVMVVVCMT SLCDPHTGQR WLPLILAGFT SGFL LLRGR SYVDRWQSIT LAGTAVIIAA AVCVRYALEL SSPLAVSIVA AILVLLPAAG MAAAAHVPHT IYSPLFRKFV EWIEY LCLM PIFPLALWLM NVYAAIRYR UniProtKB: ESX-5 secretion system protein EccD5 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 59.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)