+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10207 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | A 4.2 Angstrom structure of HIV-1 CA-SP1 | |||||||||
 Map data Map data | A 4.2A structure of HIV-1 CA-SP1. | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||
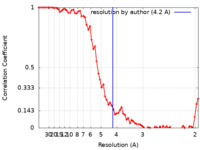
| Method | subtomogram averaging / cryo EM / Resolution: 4.2 Å | |||||||||
 Authors Authors | Turonova B / Hagen WJH / Obr M / Mosalaganti S / Kraeusslich HG / Beck M | |||||||||
| Funding support |  Germany, 1 items Germany, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Benchmarking tomographic acquisition schemes for high-resolution structural biology. Authors: Beata Turoňová / Wim J H Hagen / Martin Obr / Shyamal Mosalaganti / J Wouter Beugelink / Christian E Zimmerli / Hans-Georg Kräusslich / Martin Beck /    Abstract: Cryo electron tomography with subsequent subtomogram averaging is a powerful technique to structurally analyze macromolecular complexes in their native context. Although close to atomic resolution in ...Cryo electron tomography with subsequent subtomogram averaging is a powerful technique to structurally analyze macromolecular complexes in their native context. Although close to atomic resolution in principle can be obtained, it is not clear how individual experimental parameters contribute to the attainable resolution. Here, we have used immature HIV-1 lattice as a benchmarking sample to optimize the attainable resolution for subtomogram averaging. We systematically tested various experimental parameters such as the order of projections, different angular increments and the use of the Volta phase plate. We find that although any of the prominently used acquisition schemes is sufficient to obtain subnanometer resolution, dose-symmetric acquisition provides considerably better outcome. We discuss our findings in order to provide guidance for data acquisition. Our data is publicly available and might be used to further develop processing routines. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10277 (Title: Tilt-schemes benchmarking for cryo electron tomography of HIV-1 CA-SP1 EMPIAR-10277 (Title: Tilt-schemes benchmarking for cryo electron tomography of HIV-1 CA-SP1Data size: 45.6 Data #1: Unsorted raw tilt-series for benchmarked tilt-schemes [tilt series] Data #2: Tilt-series collected using the dose-symmetric scheme with 3 degrees tilt-step without any gold fiducials [tilt series]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10207.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10207.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | A 4.2A structure of HIV-1 CA-SP1. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3269 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
+Additional map: A CTF-reweighted odd half-map of HIV-1 CA-SP1 obtained...
+Additional map: A CTF-reweighted odd half-map of HIV-1 CA-SP1 obtained...
+Additional map: A CTF-reweighted even half-map of HIV-1 CA-SP1 obtained...
+Additional map: A CTF-reweighted odd half-map of HIV-1 CA-SP1 obtained...
+Additional map: A CTF-reweighted odd half-map of HIV-1 CA-SP1 obtained...
+Additional map: A CTF-reweighted even half-map of HIV-1 CA-SP1 obtained...
+Additional map: A CTF-reweighted map of HIV-1 CA-SP1 obtained with...
+Additional map: A CTF-reweighted odd half-map of HIV-1 CA-SP1 obtained...
+Additional map: A CTF-reweighted map of HIV-1 CA-SP1 obtained with...
+Additional map: A CTF-reweighted odd half-map of HIV-1 CA-SP1 obtained...
+Additional map: A CTF-reweighted even half-map of HIV-1 CA-SP1 obtained...
+Additional map: A CTF-reweighted even half-map of HIV-1 CA-SP1 obtained...
+Additional map: A CTF-reweighted map of HIV-1 CA-SP1 obtained with...
+Additional map: A CTF-reweighted odd half-map of HIV-1 CA-SP1 obtained...
+Additional map: A CTF-reweighted even half-map of HIV-1 CA-SP1 obtained...
+Additional map: A CTF-reweighted odd half-map of HIV-1 CA-SP1 obtained...
+Additional map: A CTF-reweighted even half-map of HIV-1 CA-SP1 obtained...
+Additional map: A CTF-reweighted map of HIV-1 CA-SP1 obtained with...
+Additional map: A raw map of HIV-1 CA-SP1 obtained with...
+Additional map: A raw odd half-map of HIV-1 CA-SP1 obtained...
+Additional map: A raw even half-map of HIV-1 CA-SP1 obtained...
+Additional map: A raw map of HIV-1 CA-SP1 obtained with...
+Additional map: A CTF-reweighted odd half-map of HIV-1 CA-SP1 obtained...
+Additional map: A raw odd half-map of HIV-1 CA-SP1 obtained...
+Additional map: A raw even half-map of HIV-1 CA-SP1 obtained...
+Additional map: A raw map of HIV-1 CA-SP1 obtained with...
+Additional map: A raw odd half-map of HIV-1 CA-SP1 obtained...
+Additional map: A raw even half-map of HIV-1 CA-SP1 obtained...
+Additional map: A CTF-reweighted even half-map of HIV-1 CA-SP1 obtained...
+Additional map: A raw even half-map of HIV-1 CA-SP1 obtained...
+Additional map: A raw map of HIV-1 CA-SP1 obtained with...
+Additional map: A raw map of HIV-1 CA-SP1 obtained with...
+Additional map: A CTF-reweighted map of HIV-1 CA-SP1 obtained with...
+Additional map: A CTF-reweighted even half-map of HIV-1 CA-SP1 obtained...
+Additional map: A CTF-reweighted odd half-map of HIV-1 CA-SP1 obtained...
+Additional map: A CTF-reweighted even half-map of HIV-1 CA-SP1 obtained...
+Additional map: A CTF-reweighted even half-map of HIV-1 CA-SP1 obtained...
+Additional map: A CTF-reweighted odd half-map of HIV-1 CA-SP1 obtained...
+Additional map: A CTF-reweighted even half-map of HIV-1 CA-SP1 obtained...
- Sample components
Sample components
-Entire : Human immunodeficiency virus 1
| Entire | Name:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
|---|---|
| Components |
|
-Supramolecule #1: Human immunodeficiency virus 1
| Supramolecule | Name: Human immunodeficiency virus 1 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 11676 / Sci species name: Human immunodeficiency virus 1 / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: OTHER / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Host system | Organism:  |
-Macromolecule #1: Virus-like particle
| Macromolecule | Name: Virus-like particle / type: other / ID: 1 / Classification: other |
|---|---|
| Sequence | String: Not applic able |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 288.15 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 3708 pixel / Digitization - Dimensions - Height: 3838 pixel / Number grids imaged: 1 / Average electron dose: 3.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 4.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)