[English] 日本語
 Yorodumi
Yorodumi- EMDB-12518: Structure of the periplasmic assembly from the ESX-5 inner membra... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12518 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
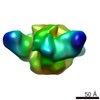
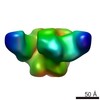
| Title | Structure of the periplasmic assembly from the ESX-5 inner membrane complex, C1 symmetry | |||||||||||||||
 Map data Map data | -50 Bfactor sharpening of 3D refinement map. | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | T7SS / mycobacteria / protein transport / secretion / type VII secretion system / membrane / MEMBRANE PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationHydrolases; Acting on acid anhydrides / Hydrolases; Acting on peptide bonds (peptidases); Serine endopeptidases / peptidoglycan-based cell wall / protein processing / serine-type endopeptidase activity / hydrolase activity / ATP binding / plasma membrane Similarity search - Function | |||||||||||||||
| Biological species |  Mycobacterium tuberculosis H37Rv (bacteria) / Mycobacterium tuberculosis H37Rv (bacteria) /  Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) | |||||||||||||||
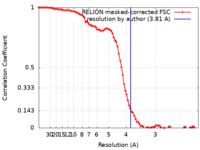
| Method | single particle reconstruction / cryo EM / Resolution: 3.81 Å | |||||||||||||||
 Authors Authors | Bunduc CM / Fahrenkamp D / Wald J / Ummels R / Bitter W / Houben ENG / Marlovits TC | |||||||||||||||
| Funding support |  Germany, Germany,  Netherlands, European Union, 4 items Netherlands, European Union, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: Structure and dynamics of a mycobacterial type VII secretion system. Authors: Catalin M Bunduc / Dirk Fahrenkamp / Jiri Wald / Roy Ummels / Wilbert Bitter / Edith N G Houben / Thomas C Marlovits /   Abstract: Mycobacterium tuberculosis is the cause of one of the most important infectious diseases in humans, which leads to 1.4 million deaths every year. Specialized protein transport systems-known as ...Mycobacterium tuberculosis is the cause of one of the most important infectious diseases in humans, which leads to 1.4 million deaths every year. Specialized protein transport systems-known as type VII secretion systems (T7SSs)-are central to the virulence of this pathogen, and are also crucial for nutrient and metabolite transport across the mycobacterial cell envelope. Here we present the structure of an intact T7SS inner-membrane complex of M. tuberculosis. We show how the 2.32-MDa ESX-5 assembly, which contains 165 transmembrane helices, is restructured and stabilized as a trimer of dimers by the MycP protease. A trimer of MycP caps a central periplasmic dome-like chamber that is formed by three EccB dimers, with the proteolytic sites of MycP facing towards the cavity. This chamber suggests a central secretion and processing conduit. Complexes without MycP show disruption of the EccB periplasmic assembly and increased flexibility, which highlights the importance of MycP for complex integrity. Beneath the EccB-MycP chamber, dimers of the EccC ATPase assemble into three bundles of four transmembrane helices each, which together seal the potential central secretion channel. Individual cytoplasmic EccC domains adopt two distinctive conformations that probably reflect different secretion states. Our work suggests a previously undescribed mechanism of protein transport and provides a structural scaffold to aid in the development of drugs against this major human pathogen. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12518.map.gz emd_12518.map.gz | 5.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12518-v30.xml emd-12518-v30.xml emd-12518.xml emd-12518.xml | 19.1 KB 19.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12518_fsc.xml emd_12518_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_12518.png emd_12518.png | 93.7 KB | ||
| Masks |  emd_12518_msk_1.map emd_12518_msk_1.map | 59.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-12518.cif.gz emd-12518.cif.gz | 6.1 KB | ||
| Others |  emd_12518_additional_1.map.gz emd_12518_additional_1.map.gz emd_12518_half_map_1.map.gz emd_12518_half_map_1.map.gz emd_12518_half_map_2.map.gz emd_12518_half_map_2.map.gz | 45.6 MB 45.9 MB 45.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12518 http://ftp.pdbj.org/pub/emdb/structures/EMD-12518 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12518 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12518 | HTTPS FTP |
-Related structure data
| Related structure data |  7npsMC  7np7C  7nprC  7nptC  7npuC  7npvC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12518.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12518.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | -50 Bfactor sharpening of 3D refinement map. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_12518_msk_1.map emd_12518_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: 3D refinement map.
| File | emd_12518_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D refinement map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half1 of 3D refinement map.
| File | emd_12518_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half1 of 3D refinement map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half2 of 3D refinement map.
| File | emd_12518_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half2 of 3D refinement map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Periplasmic assembly of the ESX-5 inner membrane complex
| Entire | Name: Periplasmic assembly of the ESX-5 inner membrane complex |
|---|---|
| Components |
|
-Supramolecule #1: Periplasmic assembly of the ESX-5 inner membrane complex
| Supramolecule | Name: Periplasmic assembly of the ESX-5 inner membrane complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis H37Rv (bacteria) Mycobacterium tuberculosis H37Rv (bacteria) |
-Macromolecule #1: ESX-5 secretion system ATPase EccB5
| Macromolecule | Name: ESX-5 secretion system ATPase EccB5 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: Hydrolases; Acting on acid anhydrides |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria)Strain: ATCC 25618 / H37Rv |
| Molecular weight | Theoretical: 53.769988 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Sequence | String: MAEESRGQRG SGYGLGLSTR TQVTGYQFLA RRTAMALTRW RVRMEIEPGR RQTLAVVASV SAALVICLGA LLWSFISPSG QLNESPIIA DRDSGALYVR VGDRLYPALN LASARLITGR PDNPHLVRSS QIATMPRGPL VGIPGAPSSF SPKSPPASSW L VCDTVATS ...String: MAEESRGQRG SGYGLGLSTR TQVTGYQFLA RRTAMALTRW RVRMEIEPGR RQTLAVVASV SAALVICLGA LLWSFISPSG QLNESPIIA DRDSGALYVR VGDRLYPALN LASARLITGR PDNPHLVRSS QIATMPRGPL VGIPGAPSSF SPKSPPASSW L VCDTVATS SSIGSLQGVT VTVIDGTPDL TGHRQILSGS DAVVLRYGGD AWVIREGRRS RIEPTNRAVL LPLGLTPEQV SQ ARPMSRA LFDALPVGPE LLVPEVPNAG GPATFPGAPG PIGTVIVTPQ ISGPQQYSLV LGDGVQTLPP LVAQILQNAG SAG NTKPLT VEPSTLAKMP VVNRLDLSAY PDNPLEVVDI REHPSTCWWW ERTAGENRAR VRVVSGPTIP VAATEMNKVV SLVK ADTSG RQADQVYFGP DHANFVAVTG NNPGAQTSES LWWVTDAGAR FGVEDSKEAR DALGLTLTPS LAPWVALRLL PQGPT LSRA DALVEHDTLP MDMTPAELVV PK UniProtKB: ESX-5 secretion system ATPase EccB5 |
-Macromolecule #2: Mycosin-5
| Macromolecule | Name: Mycosin-5 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO EC number: Hydrolases; Acting on peptide bonds (peptidases); Serine endopeptidases |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria) Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) (bacteria)Strain: ATCC 25618 / H37Rv |
| Molecular weight | Theoretical: 60.074211 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Sequence | String: MQRFGTGSSR SWCGRAGTAT IAAVLLASGA LTGLPPAYAI SPPTIDPGAL PPDGPPGPLA PMKQNAYCTE VGVLPGTDFQ LQPKYMEML NLNEAWQFGR GDGVKVAVID TGVTPHPRLP RLIPGGDYVM AGGDGLSDCD AHGTLVASMI AAVPANGAVP L PSVPRRPV ...String: MQRFGTGSSR SWCGRAGTAT IAAVLLASGA LTGLPPAYAI SPPTIDPGAL PPDGPPGPLA PMKQNAYCTE VGVLPGTDFQ LQPKYMEML NLNEAWQFGR GDGVKVAVID TGVTPHPRLP RLIPGGDYVM AGGDGLSDCD AHGTLVASMI AAVPANGAVP L PSVPRRPV TIPTTETPPP PQTVTLSPVP PQTVTVIPAP PPEEGVPPGA PVPGPEPPPA PGPQPPAVDR GGGTVTVPSY SG GRKIAPI DNPRNPHPSA PSPALGPPPD AFSGIAPGVE IISIRQSSQA FGLKDPYTGD EDPQTAQKID NVETMARAIV HAA NMGASV INISDVMCMS ARNVIDQRAL GAAVHYAAVD KDAVIVAAAG DGSKKDCKQN PIFDPLQPDD PRAWNAVTTV VTPS WFHDY VLTVGAVDAN GQPLSKMSIA GPWVSISAPG TDVVGLSPRD DGLINAIDGP DNSLLVPAGT SFSAAIVSGV AALVR AKFP ELSAYQIINR LIHTARPPAR GVDNQVGYGV VDPVAALTWD VPKGPAEPPK QLSAPLVVPQ PPAPRDMVPI WVAAGG LAG ALLIGGAVFG TATLMRRSRK QQ UniProtKB: Mycosin-5 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE-PROPANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 59.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)