[English] 日本語
 Yorodumi
Yorodumi- EMDB-12357: ordered protein arrays on the axoneme-facing surface of the outer... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12357 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | ordered protein arrays on the axoneme-facing surface of the outer mitochondrial membrane in pig sperm mitochondria (alignment focused on one unit) | |||||||||
 Map data Map data | subtomogram average of ordered protein arrays on the axoneme-facing surface of the outer mitochondrial membrane in pig sperm mitochondria (alignment focused on one unit) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | mitochondria / sperm / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationPINK1-PRKN Mediated Mitophagy / Triglyceride biosynthesis / Ub-specific processing proteases / glycerol-3-phosphate biosynthetic process / voltage-gated monoatomic anion channel activity / mitochondrial outer membrane permeabilization / glycerol kinase / glycerol kinase activity / glycerol metabolic process / ceramide binding ...PINK1-PRKN Mediated Mitophagy / Triglyceride biosynthesis / Ub-specific processing proteases / glycerol-3-phosphate biosynthetic process / voltage-gated monoatomic anion channel activity / mitochondrial outer membrane permeabilization / glycerol kinase / glycerol kinase activity / glycerol metabolic process / ceramide binding / voltage-gated monoatomic ion channel activity / phosphatidylcholine binding / oxysterol binding / glycerol catabolic process / monoatomic anion transport / triglyceride metabolic process / cholesterol binding / lipid transport / porin activity / pore complex / sperm midpiece / mitochondrial outer membrane / nucleotide binding / mitochondrion / ATP binding / membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 35.0 Å | |||||||||
 Authors Authors | Leung MR / Zeev-Ben-Mordehai T | |||||||||
| Funding support |  Netherlands, 1 items Netherlands, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2021 Journal: Proc Natl Acad Sci U S A / Year: 2021Title: In-cell structures of conserved supramolecular protein arrays at the mitochondria-cytoskeleton interface in mammalian sperm. Authors: Miguel Ricardo Leung / Riccardo Zenezini Chiozzi / Marc C Roelofs / Johannes F Hevler / Ravi Teja Ravi / Paula Maitan / Min Zhang / Heiko Henning / Elizabeth G Bromfield / Stuart C Howes / ...Authors: Miguel Ricardo Leung / Riccardo Zenezini Chiozzi / Marc C Roelofs / Johannes F Hevler / Ravi Teja Ravi / Paula Maitan / Min Zhang / Heiko Henning / Elizabeth G Bromfield / Stuart C Howes / Bart M Gadella / Albert J R Heck / Tzviya Zeev-Ben-Mordehai /     Abstract: Mitochondria-cytoskeleton interactions modulate cellular physiology by regulating mitochondrial transport, positioning, and immobilization. However, there is very little structural information ...Mitochondria-cytoskeleton interactions modulate cellular physiology by regulating mitochondrial transport, positioning, and immobilization. However, there is very little structural information defining mitochondria-cytoskeleton interfaces in any cell type. Here, we use cryofocused ion beam milling-enabled cryoelectron tomography to image mammalian sperm, where mitochondria wrap around the flagellar cytoskeleton. We find that mitochondria are tethered to their neighbors through intermitochondrial linkers and are anchored to the cytoskeleton through ordered arrays on the outer mitochondrial membrane. We use subtomogram averaging to resolve in-cell structures of these arrays from three mammalian species, revealing they are conserved across species despite variations in mitochondrial dimensions and cristae organization. We find that the arrays consist of boat-shaped particles anchored on a network of membrane pores whose arrangement and dimensions are consistent with voltage-dependent anion channels. Proteomics and in-cell cross-linking mass spectrometry suggest that the conserved arrays are composed of glycerol kinase-like proteins. Ordered supramolecular assemblies may serve to stabilize similar contact sites in other cell types in which mitochondria need to be immobilized in specific subcellular environments, such as in muscles and neurons. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12357.map.gz emd_12357.map.gz | 454.4 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12357-v30.xml emd-12357-v30.xml emd-12357.xml emd-12357.xml | 13.7 KB 13.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_12357.png emd_12357.png | 152.5 KB | ||
| Filedesc metadata |  emd-12357.cif.gz emd-12357.cif.gz | 5.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12357 http://ftp.pdbj.org/pub/emdb/structures/EMD-12357 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12357 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12357 | HTTPS FTP |
-Related structure data
| Related structure data |  7nieMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_12357.map.gz / Format: CCP4 / Size: 501 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12357.map.gz / Format: CCP4 / Size: 501 KB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
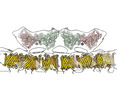
| Annotation | subtomogram average of ordered protein arrays on the axoneme-facing surface of the outer mitochondrial membrane in pig sperm mitochondria (alignment focused on one unit) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4.34 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
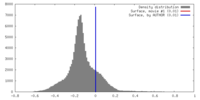
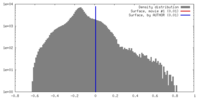
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : subtomogram average of ordered protein arrays on the axoneme-faci...
| Entire | Name: subtomogram average of ordered protein arrays on the axoneme-facing surface of the outer mitochondrial membrane in pig sperm mitochondria (alignment focused on one unit) |
|---|---|
| Components |
|
-Supramolecule #1: subtomogram average of ordered protein arrays on the axoneme-faci...
| Supramolecule | Name: subtomogram average of ordered protein arrays on the axoneme-facing surface of the outer mitochondrial membrane in pig sperm mitochondria (alignment focused on one unit) type: cell / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Glycerol kinase
| Macromolecule | Name: Glycerol kinase / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO / EC number: glycerol kinase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 53.579547 KDa |
| Sequence | String: MAASKKAVLG PLVGAVDQGT SSTRFLVFNS KTAELLSHHQ VEIEQKFPKE GWVEQDPKEI LQSVYECIEK TCEKLGQLNI DISNIKAIG VSNQRETTVV WDKLTGEPLY NAVVWLDLRT QSTVANLSKI IPVNNNFVKT KTGLPLSTYF SAVKLRWLLD N VRKIQKAV ...String: MAASKKAVLG PLVGAVDQGT SSTRFLVFNS KTAELLSHHQ VEIEQKFPKE GWVEQDPKEI LQSVYECIEK TCEKLGQLNI DISNIKAIG VSNQRETTVV WDKLTGEPLY NAVVWLDLRT QSTVANLSKI IPVNNNFVKT KTGLPLSTYF SAVKLRWLLD N VRKIQKAV EEDRALFGTI DSWLIWSLTG GARGGVHCTD VTNASRTMLF NIHSLEWDKE LCEFFEVPMK ILPNVRSSSE IY GLMKAGA LEGVPISGCL GDQSAALVGQ MCFQDGQAKN TYGTGCFLLC NTGHKCSVAI AGAVVRWLRD NLGIIKTSEE IEK LAKEVG TSYGCYFVPA FSGLYAPYWE PSARGIICGL TQFTNKCHIA FAALEAVCFQ TREILDAMNR DCGIPLSHLQ VDGG MTNNK ILMQLQADIL YIPVVKPSMP ETTALGAAMA AGAAEGVGVW SLKPEDLSAV TMERFEPQIN AKESEIRYST WKKAV KKSM GWV UniProtKB: glycerol kinase |
-Macromolecule #2: Voltage-dependent anion-selective channel protein 2
| Macromolecule | Name: Voltage-dependent anion-selective channel protein 2 / type: protein_or_peptide / ID: 2 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 31.637484 KDa |
| Sequence | String: MASYGQTCPR PMCIPPSYAD LGKAARDIFN KGFGFGLVKL DVKTKSCSGV EFSTSGSSNT DTGKVTGTLE TKYKWCEYGL TFTEKWNTD NTLGTEIAIE DQICQGLKLT FDTTFSPNTG KKSGKIKSSY KRECVNLGCD VDFDFAGPAI HGSAVFGYEG W LAGYQMTF ...String: MASYGQTCPR PMCIPPSYAD LGKAARDIFN KGFGFGLVKL DVKTKSCSGV EFSTSGSSNT DTGKVTGTLE TKYKWCEYGL TFTEKWNTD NTLGTEIAIE DQICQGLKLT FDTTFSPNTG KKSGKIKSSY KRECVNLGCD VDFDFAGPAI HGSAVFGYEG W LAGYQMTF DSAKSKLTRN NFAVGYRTGD FQLHTNVNDG TEFGGSIYQK VCEDLDTSVN LAWTSGTNCT RFGIAAKYQL DP TASISAK VNNSSLIGVG YTQTLRPGVK LTLSALVDGK SINAGGHKLG LALELEA UniProtKB: Non-selective voltage-gated ion channel VDAC2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE-PROPANE / Instrument: HOMEMADE PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 1.3 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Number classes used: 1 / Applied symmetry - Point group: C2 (2 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 35.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: PEET (ver. 1.13.0) / Number subtomograms used: 536 |
|---|---|
| Extraction | Number tomograms: 3 / Number images used: 268 / Reference model: random particle from the dataset / Software - Name: PEET (ver. 1.13.0) |
| Final angle assignment | Type: OTHER / Software - Name: PEET (ver. 1.13.0) |
-Atomic model buiding 1
| Details | Homology models of pig glycerol kinase and VDAC2 were made using Robetta. Rigid body fitting was done with Chimera/ChimeraX. Putative GK proteins only fit in one orientation, but for VDAC proteins the rotation around the pore axis remains ambiguous. |
|---|---|
| Refinement | Protocol: RIGID BODY FIT |
| Output model |  PDB-7nie: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)