+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10413 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SAGA Tra1 module | |||||||||
 Map data Map data | local resolution filtered map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Coactivator / Transcription / Histone acetyltransferase / Histone deubiquitinase / GENE REGULATION | |||||||||
| Function / homology |  Function and homology information Function and homology informationTTT Hsp90 cochaperone complex / SLIK (SAGA-like) complex / SAGA complex / RNA Polymerase II Promoter Escape / RNA Polymerase II Transcription Pre-Initiation And Promoter Opening / RNA Polymerase II Transcription Initiation / RNA Polymerase II Transcription Initiation And Promoter Clearance / IRE1-mediated unfolded protein response / RNA Polymerase II Pre-transcription Events / transcription factor TFIID complex ...TTT Hsp90 cochaperone complex / SLIK (SAGA-like) complex / SAGA complex / RNA Polymerase II Promoter Escape / RNA Polymerase II Transcription Pre-Initiation And Promoter Opening / RNA Polymerase II Transcription Initiation / RNA Polymerase II Transcription Initiation And Promoter Clearance / IRE1-mediated unfolded protein response / RNA Polymerase II Pre-transcription Events / transcription factor TFIID complex / DNA repair-dependent chromatin remodeling / NuA4 histone acetyltransferase complex / Ub-specific processing proteases / RNA polymerase II preinitiation complex assembly / TBP-class protein binding / transcription coregulator activity / peroxisome / chromatin organization / transcription by RNA polymerase II / molecular adaptor activity / RNA polymerase II-specific DNA-binding transcription factor binding / protein heterodimerization activity / DNA repair / chromatin binding / regulation of transcription by RNA polymerase II / regulation of DNA-templated transcription / DNA-templated transcription / positive regulation of transcription by RNA polymerase II / DNA binding / identical protein binding / nucleus Similarity search - Function | |||||||||
| Biological species |  | |||||||||
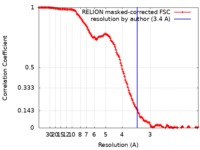
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Wang H / Cheung A | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2020 Journal: Nature / Year: 2020Title: Structure of the transcription coactivator SAGA. Authors: Haibo Wang / Christian Dienemann / Alexandra Stützer / Henning Urlaub / Alan C M Cheung / Patrick Cramer /   Abstract: Gene transcription by RNA polymerase II is regulated by activator proteins that recruit the coactivator complexes SAGA (Spt-Ada-Gcn5-acetyltransferase) and transcription factor IID (TFIID). SAGA is ...Gene transcription by RNA polymerase II is regulated by activator proteins that recruit the coactivator complexes SAGA (Spt-Ada-Gcn5-acetyltransferase) and transcription factor IID (TFIID). SAGA is required for all regulated transcription and is conserved among eukaryotes. SAGA contains four modules: the activator-binding Tra1 module, the core module, the histone acetyltransferase (HAT) module and the histone deubiquitination (DUB) module. Previous studies provided partial structures, but the structure of the central core module is unknown. Here we present the cryo-electron microscopy structure of SAGA from the yeast Saccharomyces cerevisiae and resolve the core module at 3.3 Å resolution. The core module consists of subunits Taf5, Sgf73 and Spt20, and a histone octamer-like fold. The octamer-like fold comprises the heterodimers Taf6-Taf9, Taf10-Spt7 and Taf12-Ada1, and two histone-fold domains in Spt3. Spt3 and the adjacent subunit Spt8 interact with the TATA box-binding protein (TBP). The octamer-like fold and its TBP-interacting region are similar in TFIID, whereas Taf5 and the Taf6 HEAT domain adopt distinct conformations. Taf12 and Spt20 form flexible connections to the Tra1 module, whereas Sgf73 tethers the DUB module. Binding of a nucleosome to SAGA displaces the HAT and DUB modules from the core-module surface, allowing the DUB module to bind one face of an ubiquitinated nucleosome. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10413.map.gz emd_10413.map.gz | 142.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10413-v30.xml emd-10413-v30.xml emd-10413.xml emd-10413.xml | 23.2 KB 23.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10413_fsc.xml emd_10413_fsc.xml | 14.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_10413.png emd_10413.png | 302 KB | ||
| Filedesc metadata |  emd-10413.cif.gz emd-10413.cif.gz | 8.7 KB | ||
| Others |  emd_10413_half_map_1.map.gz emd_10413_half_map_1.map.gz emd_10413_half_map_2.map.gz emd_10413_half_map_2.map.gz | 194.1 MB 194.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10413 http://ftp.pdbj.org/pub/emdb/structures/EMD-10413 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10413 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10413 | HTTPS FTP |
-Related structure data
| Related structure data |  6t9jMC  6t9iC  6t9kC  6t9lC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10413.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10413.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | local resolution filtered map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
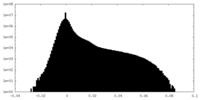
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
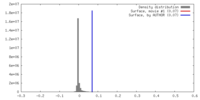
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: half map 1
| File | emd_10413_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
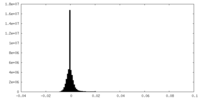
| Density Histograms |
-Half map: half map 2
| File | emd_10413_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
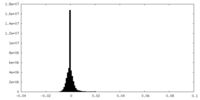
| Density Histograms |
- Sample components
Sample components
-Entire : SAGA Tra1 module
| Entire | Name: SAGA Tra1 module |
|---|---|
| Components |
|
-Supramolecule #1: SAGA Tra1 module
| Supramolecule | Name: SAGA Tra1 module / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 450 KDa |
-Macromolecule #1: Transcription factor SPT20
| Macromolecule | Name: Transcription factor SPT20 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 67.880312 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSANSPTGND PHVFGIPVNA TPSNMGSPGS PVNVPPPMNP AVANVNHPVM RTNSNSNANE GTRTLTREQI QQLQQRQRLL LQQRLLEQQ RKQQALQNYE AQFYQMLMTL NKRPKRLYNF VEDADSILKK YEQYLHSFEF HIYENNYKIC APANSRLQQQ Q KQPELTSD ...String: MSANSPTGND PHVFGIPVNA TPSNMGSPGS PVNVPPPMNP AVANVNHPVM RTNSNSNANE GTRTLTREQI QQLQQRQRLL LQQRLLEQQ RKQQALQNYE AQFYQMLMTL NKRPKRLYNF VEDADSILKK YEQYLHSFEF HIYENNYKIC APANSRLQQQ Q KQPELTSD GLILTKNNET LKEFLEYVAR GRIPDAIMEV LRDCNIQFYE GNLILQVYDH TNTVDVTPKE NKPNLNSSSS PS NNNSTQD NSKIQQPSEP NSGVANTGAN TANKKASFKR PRVYRTLLKP NDLTTYYDMM SYADNARFSD SIYQQFESEI LTL TKRNLS LSVPLNPYEH RDMLEETAFS EPHWDSEKKS FIHEHRAEST REGTKGVVGH IEERDEFPQH SSNYEQLMLI MNER TTTIT NSTFAVSLTK NAMEIASSSS NGVRGASSST SNSASNTRNN SLANGNQVAL AAAAAAAAVG STMGNDNNQF SRLKF IEQW RINKEKRKQQ ALSANINPTP FNARISMTAP LTPQQQLLQR QQQALEQQQN GGAMKNANKR SGNNATSNNN NNNNNL DKP KVKRPRKNAK KSESGTPAPK KKRMTKKKQS ASSTPSSTTM S UniProtKB: SAGA complex subunit SPT20 |
-Macromolecule #2: Transcription initiation factor TFIID subunit 12
| Macromolecule | Name: Transcription initiation factor TFIID subunit 12 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 61.144379 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSSNPENSGV NANNNTGTGN ADAITGAQQN MVLQPRQLQE MAAKFRTLLT EARNVGETTP RGKELMFQAA KIKQVYDALT LNRRRQQAA QAYNNTSNSN SSNPASIPTE NVPNSSQQQQ QQQQQTRNNS NKFSNMIKQV LTPEENQEYE KLWQNFQVRH T SIKEKETY ...String: MSSNPENSGV NANNNTGTGN ADAITGAQQN MVLQPRQLQE MAAKFRTLLT EARNVGETTP RGKELMFQAA KIKQVYDALT LNRRRQQAA QAYNNTSNSN SSNPASIPTE NVPNSSQQQQ QQQQQTRNNS NKFSNMIKQV LTPEENQEYE KLWQNFQVRH T SIKEKETY LKQNIDRLEQ EINKQTDEGP KQQLQEKKIE LLNDWKVLKI EYTKLFNNYQ NSKKTFYVEC ARHNPALHKF LQ ESTQQQR VQQQRVQQQQ QQQQQQQQQQ QQQQQQQQQR QGQNQRKISS SNSTEIPSVT GPDALKSQQQ QQNTITATNN PRG NVNTSQ TEQSKAKVTN VNATASMLNN ISSSKSAIFK QTEPAIPISE NISTKTPAPV AYRSNRPTIT GGSAMNASAL NTPA TTKLP PYEMDTQRVM SKRKLRELVK TVGIDEGDGE TVIDGDVEEL LLDLADDFVT NVTAFSCRLA KHRKSDNLEA RDIQL HLER NWNIRIPGYS ADEIRSTRKW NPSQNYNQKL QSITSDKVAA AKNNGNNVAS LNTKK UniProtKB: SAGA complex/transcription factor TFIID complex subunit TAF12 |
-Macromolecule #3: Transcription-associated protein 1
| Macromolecule | Name: Transcription-associated protein 1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 433.677281 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSLTEQIEQF ASRFRDDDAT LQSRYSTLSE LYDIMELLNS PEDYHFFLQA VIPLLLNQLK EVPISYDAHS PEQKLRNSML DIFNRCLMN QTFQPYAMEV LEFLLSVLPK ENEENGILCM KVLTTLFKSF KSILQDKLDS FIRIIIQIYK NTPNLINQTF Y EAGKAEQG ...String: MSLTEQIEQF ASRFRDDDAT LQSRYSTLSE LYDIMELLNS PEDYHFFLQA VIPLLLNQLK EVPISYDAHS PEQKLRNSML DIFNRCLMN QTFQPYAMEV LEFLLSVLPK ENEENGILCM KVLTTLFKSF KSILQDKLDS FIRIIIQIYK NTPNLINQTF Y EAGKAEQG DLDSPKEPQA DELLDEFSKN DEEKDFPSKQ SSTEPRFENS TSSNGLRSSM FSFKILSECP ITMVTLYSSY KQ LTSTSLP EFTPLIMNLL NIQIKQQQEA REQAESRGEH FTSISTEIIN RPAYCDFILA QIKATSFLAY VFIRGYAPEF LQD YVNFVP DLIIRLLQDC PSELSSARKE LLHATRHILS TNYKKLFLPK LDYLFDERIL IGNGFTMHET LRPLAYSTVA DFIH NIRSE LQLSEIEKTI KIYTGYLLDE SLALTVQIMS AKLLLNLVER ILKLGKENPQ EAPRAKKLLM IIIDSYMNRF KTLNR QYDT IMKYYGRYET HKKEKAEKLK NSIQDNDKES EEFMRKVLEP SDDDHLMPQP KKEDINDSPD VEMTESDKVV KNDVEM FDI KNYAPILLLP TPTNDPIKDA FYLYRTLMSF LKTIIHDLKV FNPPPNEYTV ANPKLWASVS RVFSYEEVIV FKDLFHE CI IGLKFFKDHN EKLSPETTKK HFDISMPSLP VSATKDAREL MDYLAFMFMQ MDNATFNEII EQELPFVYER MLEDSGLL H VAQSFLTSEI TSPNFAGILL RFLKGKLKDL GNVDFNTSNV LIRLFKLSFM SVNLFPNINE VVLLPHLNDL ILNSLKYST TAEEPLVYFY LIRTLFRSIG GGRFENLYRS IKPILQVLLQ SLNQMILTAR LPHERELYVE LCITVPVRLS VLAPYLPFLM KPLVFALQQ YPDLVSQGLR TLELCIDNLT AEYFDPIIEP VIDDVSKALF NLLQPQPFNH AISHNVVRIL GKLGGRNRQF L KPPTDLTE KTELDIDAIA DFKINGMPED VPLSVTPGIQ SALNILQSYK SDIHYRKSAY KYLTCVLLLM TKSSAEFPTN YT ELLKTAV NSIKLERIGI EKNFDLEPTV NKRDYSNQEN LFLRLLESVF YATSIKELKD DAMDLLNNLL DHFCLLQVNT TLL NKRNYN GTFNIDLKNP NFMLDSSLIL DAIPFALSYY IPEVREVGVL AYKRIYEKSC LIYGEELALS HSFIPELAKQ FIHL CYDET YYNKRGGVLG IKVLIDNVKS SSVFLKKYQY NLANGLLFVL KDTQSEAPSA ITDSAEKLLI DLLSITFADV KEEDL GNKV LENTLTDIVC ELSNANPKVR NACQKSLHTI SNLTGIPIVK LMDHSKQFLL SPIFAKPLRA LPFTMQIGNV DAITFC LSL PNTFLTFNEE LFRLLQESIV LADAEDESLS TNIQKTTEYS TSEQLVQLRI ACIKLLAIAL KNEEFATAQQ GNIRIRI LA VFFKTMLKTS PEIINTTYEA LKGSLAENSK LPKELLQNGL KPLLMNLSDH QKLTVPGLDA LSKLLELLIA YFKVEIGR K LLDHLTAWCR VEVLDTLFGQ DLAEQMPTKI IVSIINIFHL LPPQADMFLN DLLLKVMLLE RKLRLQLDSP FRTPLARYL NRFHNPVTEY FKKNMTLRQL VLFMCNIVQR PEAKELAEDF EKELDNFYDF YISNIPKNQV RVVSFFTNMV DLFNTMVITN GDEWLKKKG NMILKLKDML NLTLKTIKEN SFYIDHLQLN QSIAKFQALY LRFTELSERD QNPLLLDFID FSFSNGIKAS Y SLKKFIFH NIIASSNKEK QNNFINDATL FVLSDKCLDA RIFVLKNVIN STLIYEVATS GSLKSYLVED KKPKWLELLH NK IWKNSNA ILAYDVLDHH DLFRFELLQL SAIFIKADPE IIAEIKKDII KFCWNFIKLE DTLIKQSAYL VTSYFISKFD FPI KVVTQV FVALLRSSHV EARYLVKQSL DVLTPVLHER MNAAGTPDTW INWVKRVMVE NSSSQNNILY QFLISHPDLF FNSR DLFIS NIIHHMNKIT FMSNSNSDSH TLAIDLASLI LYWENKTLEI TNVNNTKTDS DGDVVMSDSK SDINPVEADT TAIIV DANN NSPISLHLRE ACTAFLIRYV CASNHRAIET ELGLRAINIL SELISDKHWT NVNVKLVYFE KFLIFQDLDS ENILYY CMN ALDVLYVFFK NKTKEWIMEN LPTIQNLLEK CIKSDHHDVQ EALQKVLQVI MKAIKAQGVS VIIEEESPGK TFIQMLT SV ITQDLQETSS VTAGVTLAWV LFMNFPDNIV PLLTPLMKTF SKLCKDHLSI SQPKDAMALE EARITTKLLE KVLYILSL K VSLLGDSRRP FLSTVALLID HSMDQNFLRK IVNMSRSWIF NTEIFPTVKE KAAILTKMLA FEIRGEPSLS KLFYEIVLK LFDQEHFNNT EITVRMEQPF LVGTRVEDIG IRKRFMTILD NSLERDIKER LYYVIRDQNW EFIADYPWLN QALQLLYGSF NREKELSLK NIYCLSPPSI LQEYLPENAE MVTEVNDLEL SNFVKGHIAS MQGLCRIISS DFIDSLIEIF YQDPKAIHRA W VTLFPQVY KSIPKNEKYG FVRSIITLLS KPYHTRQISS RTNVINMLLD SISKIESLEL PPHLVKYLAI SYNAWYQSIN IL ESIQSNT SIDNTKIIEA NEDALLELYV NLQEEDMFYG LWRRRAKYTE TNIGLSYEQI GLWDKAQQLY EVAQVKARSG ALP YSQSEY ALWEDNWIQC AEKLQHWDVL TELAKHEGFT DLLLECGWRV ADWNSDRDAL EQSVKSVMDV PTPRRQMFKT FLAL QNFAE SRKGDQEVRK LCDEGIQLSL IKWVSLPIRY TPAHKWLLHG FQQYMEFLEA TQIYANLHTT TVQNLDSKAQ EIKRI LQAW RDRLPNTWDD VNMWNDLVTW RQHAFQVINN AYLPLIPALQ QSNSNSNINT HAYRGYHEIA WVINRFAHVA RKHNMP DVC ISQLARIYTL PNIEIQEAFL KLREQAKCHY QNMNELTTGL DVISNTNLVY FGTVQKAEFF TLKGMFLSKL RAYEEAN QA FATAVQIDLN LAKAWAQWGF FNDRRLSEEP NNISFASNAI SCYLQAAGLY KNSKIRELLC RILWLISIDD ASGMLTNA F DSFRGEIPVW YWITFIPQLL TSLSHKEANM VRHILIRIAK SYPQALHFQL RTTKEDFAVI QRQTMAVMGD KPDTNDRNG RRQPWEYLQE LNNILKTAYP LLALSLESLV AQINDRFKST TDEDLFRLIN VLLIDGTLNY NRLPFPRKNP KLPENTEKNL VKFSTTLLA PYIRPKFNAD FIDNKPDYET YIKRLRYWRR RLENKLDRAS KKENLEVLCP HLSNFHHQKF EDIEIPGQYL L NKDNNVHF IKIARFLPTV DFVRGTHSSY RRLMIRGHDG SVHSFAVQYP AVRHSRREER MFQLYRLFNK SLSKNVETRR RS IQFNLPI AIPLSPQVRI MNDSVSFTTL HEIHNEFCKK KGFDPDDIQD FMADKLNAAH DDALPAPDMT ILKVEIFNSI QTM FVPSNV LKDHFTSLFT QFEDFWLFRK QFASQYSSFV FMSYMMMINN RTPHKIHVDK TSGNVFTLEM LPSRFPYERV KPLL KNHDL SLPPDSPIFH NNEPVPFRLT PNIQSLIGDS ALEGIFAVNL FTISRALIEP DNELNTYLAL FIRDEIISWF SNLHR PIIE NPQLREMVQT NVDLIIRKVA QLGHLNSTPT VTTQFILDCI GSAVSPRNLA RTDVNFMPWF UniProtKB: SAGA complex/NuA4 acetyltransferase complex subunit TRA1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: Solution were made from stock solution | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 4 seconds before plunging. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average exposure time: 9.0 sec. / Average electron dose: 42.45 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Overall B value: 107.1 / Target criteria: Correlation coefficient |
|---|---|
| Output model |  PDB-6t9j: |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)