+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3824 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the SAGA and NuA4 coactivator subunit Tra1 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Coactivator / PIKK / SAGA / NuA4 / TRANSCRIPTION | |||||||||
| Function / homology |  Function and homology information Function and homology informationTTT Hsp90 cochaperone complex / SLIK (SAGA-like) complex / SAGA complex / DNA repair-dependent chromatin remodeling / NuA4 histone acetyltransferase complex / Ub-specific processing proteases / DNA repair / regulation of transcription by RNA polymerase II / regulation of DNA-templated transcription / DNA-templated transcription ...TTT Hsp90 cochaperone complex / SLIK (SAGA-like) complex / SAGA complex / DNA repair-dependent chromatin remodeling / NuA4 histone acetyltransferase complex / Ub-specific processing proteases / DNA repair / regulation of transcription by RNA polymerase II / regulation of DNA-templated transcription / DNA-templated transcription / positive regulation of transcription by RNA polymerase II / nucleus Similarity search - Function | |||||||||
| Biological species |  | |||||||||
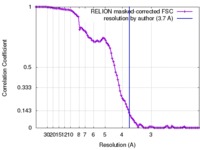
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Diaz-Santin LM / Lukoyanova N | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2017 Journal: Elife / Year: 2017Title: Cryo-EM structure of the SAGA and NuA4 coactivator subunit Tra1 at 3.7 angstrom resolution. Authors: Luis Miguel Díaz-Santín / Natasha Lukoyanova / Emir Aciyan / Alan Cm Cheung /  Abstract: Coactivator complexes SAGA and NuA4 stimulate transcription by post-translationally modifying chromatin. Both complexes contain the Tra1 subunit, a highly conserved 3744-residue protein from the ...Coactivator complexes SAGA and NuA4 stimulate transcription by post-translationally modifying chromatin. Both complexes contain the Tra1 subunit, a highly conserved 3744-residue protein from the Phosphoinositide 3-Kinase-related kinase (PIKK) family and a direct target for multiple sequence-specific activators. We present the Cryo-EM structure of Tra1 to 3.7 Å resolution, revealing an extensive network of alpha-helical solenoids organized into a diamond ring conformation and is strikingly reminiscent of DNA-PKcs, suggesting a direct role for Tra1 in DNA repair. The structure was fitted into an existing SAGA EM reconstruction and reveals limited contact surfaces to Tra1, hence it does not act as a molecular scaffold within SAGA. Mutations that affect activator targeting are distributed across the Tra1 structure, but also cluster within the N-terminal Finger region, indicating the presence of an activator interaction site. The structure of Tra1 is a key milestone in deciphering the mechanism of multiple coactivator complexes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3824.map.gz emd_3824.map.gz | 7.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3824-v30.xml emd-3824-v30.xml emd-3824.xml emd-3824.xml | 16.2 KB 16.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3824_fsc.xml emd_3824_fsc.xml | 10.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_3824.png emd_3824.png | 156.5 KB | ||
| Filedesc metadata |  emd-3824.cif.gz emd-3824.cif.gz | 8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3824 http://ftp.pdbj.org/pub/emdb/structures/EMD-3824 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3824 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3824 | HTTPS FTP |
-Related structure data
| Related structure data |  5ojsMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3824.map.gz / Format: CCP4 / Size: 93 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3824.map.gz / Format: CCP4 / Size: 93 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Tra1 - Transcription-associated protein 1
| Entire | Name: Tra1 - Transcription-associated protein 1 |
|---|---|
| Components |
|
-Supramolecule #1: Tra1 - Transcription-associated protein 1
| Supramolecule | Name: Tra1 - Transcription-associated protein 1 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 433 KDa |
-Macromolecule #1: Transcription-associated protein 1
| Macromolecule | Name: Transcription-associated protein 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 436.527281 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDYKDHDGDY KDHDIDYKDD DDKMSLTEQI EQFASRFRDD DATLQSRYST LSELYDIMEL LNSPEDYHFF LQAVIPLLLN QLKEVPISY DAHSPEQKLR NSMLDIFNRC LMNQTFQPYA MEVLEFLLSV LPKENEENGI LCMKVLTTLF KSFKSILQDK L DSFIRIII ...String: MDYKDHDGDY KDHDIDYKDD DDKMSLTEQI EQFASRFRDD DATLQSRYST LSELYDIMEL LNSPEDYHFF LQAVIPLLLN QLKEVPISY DAHSPEQKLR NSMLDIFNRC LMNQTFQPYA MEVLEFLLSV LPKENEENGI LCMKVLTTLF KSFKSILQDK L DSFIRIII QIYKNTPNLI NQTFYEAGKA EQGDLDSPKE PQADELLDEF SKNDEEKDFP SKQSSTEPRF ENSTSSNGLR SS MFSFKIL SECPITMVTL YSSYKQLTST SLPEFTPLIM NLLNIQIKQQ QEAREQAESR GEHFTSISTE IINRPAYCDF ILA QIKATS FLAYVFIRGY APEFLQDYVN FVPDLIIRLL QDCPSELSSA RKELLHATRH ILSTNYKKLF LPKLDYLFDE RILI GNGFT MHETLRPLAY STVADFIHNI RSELQLSEIE KTIKIYTGYL LDESLALTVQ IMSAKLLLNL VERILKLGKE NPQEA PRAK KLLMIIIDSY MNRFKTLNRQ YDTIMKYYGR YETHKKEKAE KLKNSIQDND KESEEFMRKV LEPSDDDHLM PQPKKE DIN DSPDVEMTES DKVVKNDVEM FDIKNYAPIL LLPTPTNDPI KDAFYLYRTL MSFLKTIIHD LKVFNPPPNE YTVANPK LW ASVSRVFSYE EVIVFKDLFH ECIIGLKFFK DHNEKLSPET TKKHFDISMP SLPVSATKDA RELMDYLAFM FMQMDNAT F NEIIEQELPF VYERMLEDSG LLHVAQSFLT SEITSPNFAG ILLRFLKGKL KDLGNVDFNT SNVLIRLFKL SFMSVNLFP NINEVVLLPH LNDLILNSLK YSTTAEEPLV YFYLIRTLFR SIGGGRFENL YRSIKPILQV LLQSLNQMIL TARLPHEREL YVELCITVP VRLSVLAPYL PFLMKPLVFA LQQYPDLVSQ GLRTLELCID NLTAEYFDPI IEPVIDDVSK ALFNLLQPQP F NHAISHNV VRILGKLGGR NRQFLKPPTD LTEKTELDID AIADFKINGM PEDVPLSVTP GIQSALNILQ SYKSDIHYRK SA YKYLTCV LLLMTKSSAE FPTNYTELLK TAVNSIKLER IGIEKNFDLE PTVNKRDYSN QENLFLRLLE SVFYATSIKE LKD DAMDLL NNLLDHFCLL QVNTTLLNKR NYNGTFNIDL KNPNFMLDSS LILDAIPFAL SYYIPEVREV GVLAYKRIYE KSCL IYGEE LALSHSFIPE LAKQFIHLCY DETYYNKRGG VLGIKVLIDN VKSSSVFLKK YQYNLANGLL FVLKDTQSEA PSAIT DSAE KLLIDLLSIT FADVKEEDLG NKVLENTLTD IVCELSNANP KVRNACQKSL HTISNLTGIP IVKLMDHSKQ FLLSPI FAK PLRALPFTMQ IGNVDAITFC LSLPNTFLTF NEELFRLLQE SIVLADAEDE SLSTNIQKTT EYSTSEQLVQ LRIACIK LL AIALKNEEFA TAQQGNIRIR ILAVFFKTML KTSPEIINTT YEALKGSLAE NSKLPKELLQ NGLKPLLMNL SDHQKLTV P GLDALSKLLE LLIAYFKVEI GRKLLDHLTA WCRVEVLDTL FGQDLAEQMP TKIIVSIINI FHLLPPQADM FLNDLLLKV MLLERKLRLQ LDSPFRTPLA RYLNRFHNPV TEYFKKNMTL RQLVLFMCNI VQRPEAKELA EDFEKELDNF YDFYISNIPK NQVRVVSFF TNMVDLFNTM VITNGDEWLK KKGNMILKLK DMLNLTLKTI KENSFYIDHL QLNQSIAKFQ ALYLRFTELS E RDQNPLLL DFIDFSFSNG IKASYSLKKF IFHNIIASSN KEKQNNFIND ATLFVLSDKC LDARIFVLKN VINSTLIYEV AT SGSLKSY LVEDKKPKWL ELLHNKIWKN SNAILAYDVL DHHDLFRFEL LQLSAIFIKA DPEIIAEIKK DIIKFCWNFI KLE DTLIKQ SAYLVTSYFI SKFDFPIKVV TQVFVALLRS SHVEARYLVK QSLDVLTPVL HERMNAAGTP DTWINWVKRV MVEN SSSQN NILYQFLISH PDLFFNSRDL FISNIIHHMN KITFMSNSNS DSHTLAIDLA SLILYWENKT LEITNVNNTK TDSDG DVVM SDSKSDINPV EADTTAIIVD ANNNSPISLH LREACTAFLI RYVCASNHRA IETELGLRAI NILSELISDK HWTNVN VKL VYFEKFLIFQ DLDSENILYY CMNALDVLYV FFKNKTKEWI MENLPTIQNL LEKCIKSDHH DVQEALQKVL QVIMKAI KA QGVSVIIEEE SPGKTFIQML TSVITQDLQE TSSVTAGVTL AWVLFMNFPD NIVPLLTPLM KTFSKLCKDH LSISQPKD A MALEEARITT KLLEKVLYIL SLKVSLLGDS RRPFLSTVAL LIDHSMDQNF LRKIVNMSRS WIFNTEIFPT VKEKAAILT KMLAFEIRGE PSLSKLFYEI VLKLFDQEHF NNTEITVRME QPFLVGTRVE DIGIRKRFMT ILDNSLERDI KERLYYVIRD QNWEFIADY PWLNQALQLL YGSFNREKEL SLKNIYCLSP PSILQEYLPE NAEMVTEVND LELSNFVKGH IASMQGLCRI I SSDFIDSL IEIFYQDPKA IHRAWVTLFP QVYKSIPKNE KYGFVRSIIT LLSKPYHTRQ ISSRTNVINM LLDSISKIES LE LPPHLVK YLAISYNAWY QSINILESIQ SNTSIDNTKI IEANEDALLE LYVNLQEEDM FYGLWRRRAK YTETNIGLSY EQI GLWDKA QQLYEVAQVK ARSGALPYSQ SEYALWEDNW IQCAEKLQHW DVLTELAKHE GFTDLLLECG WRVADWNSDR DALE QSVKS VMDVPTPRRQ MFKTFLALQN FAESRKGDQE VRKLCDEGIQ LSLIKWVSLP IRYTPAHKWL LHGFQQYMEF LEATQ IYAN LHTTTVQNLD SKAQEIKRIL QAWRDRLPNT WDDVNMWNDL VTWRQHAFQV INNAYLPLIP ALQQSNSNSN INTHAY RGY HEIAWVINRF AHVARKHNMP DVCISQLARI YTLPNIEIQE AFLKLREQAK CHYQNMNELT TGLDVISNTN LVYFGTV QK AEFFTLKGMF LSKLRAYEEA NQAFATAVQI DLNLAKAWAQ WGFFNDRRLS EEPNNISFAS NAISCYLQAA GLYKNSKI R ELLCRILWLI SIDDASGMLT NAFDSFRGEI PVWYWITFIP QLLTSLSHKE ANMVRHILIR IAKSYPQALH FQLRTTKED FAVIQRQTMA VMGDKPDTND RNGRRQPWEY LQELNNILKT AYPLLALSLE SLVAQINDRF KSTTDEDLFR LINVLLIDGT LNYNRLPFP RKNPKLPENT EKNLVKFSTT LLAPYIRPKF NADFIDNKPD YETYIKRLRY WRRRLENKLD RASKKENLEV L CPHLSNFH HQKFEDIEIP GQYLLNKDNN VHFIKIARFL PTVDFVRGTH SSYRRLMIRG HDGSVHSFAV QYPAVRHSRR EE RMFQLYR LFNKSLSKNV ETRRRSIQFN LPIAIPLSPQ VRIMNDSVSF TTLHEIHNEF CKKKGFDPDD IQDFMADKLN AAH DDALPA PDMTILKVEI FNSIQTMFVP SNVLKDHFTS LFTQFEDFWL FRKQFASQYS SFVFMSYMMM INNRTPHKIH VDKT SGNVF TLEMLPSRFP YERVKPLLKN HDLSLPPDSP IFHNNEPVPF RLTPNIQSLI GDSALEGIFA VNLFTISRAL IEPDN ELNT YLALFIRDEI ISWFSNLHRP IIENPQLREM VQTNVDLIIR KVAQLGHLNS TPTVTTQFIL DCIGSAVSPR NLARTD VNF MPWF UniProtKB: SAGA complex/NuA4 acetyltransferase complex subunit TRA1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||||||||
| Grid | Model: Agar Scientific / Support film - Material: CARBON / Support film - topology: LACEY / Pretreatment - Type: GLOW DISCHARGE | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 94 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: Two subsequent applications of protein were required to achieve the desired particle density on grids. Each application was followed by 20 sec waiting time, with a short 0.5 sec blotting ...Details: Two subsequent applications of protein were required to achieve the desired particle density on grids. Each application was followed by 20 sec waiting time, with a short 0.5 sec blotting after first application and 5 sec blotting after the second.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 1.4 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-5ojs: |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)