+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9599 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of mouse heavy-chain apoferritin at 1.62 A | |||||||||
 Map data Map data | mouse heavy chain apoferritin sharpened map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 1.62 Å | |||||||||
 Authors Authors | Danev R / Yanagisawa H / Kikkawa M | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: Trends Biochem Sci / Year: 2019 Journal: Trends Biochem Sci / Year: 2019Title: Cryo-Electron Microscopy Methodology: Current Aspects and Future Directions. Authors: Radostin Danev / Haruaki Yanagisawa / Masahide Kikkawa /  Abstract: Cryo-electron microscopy (cryo-EM) has emerged as a powerful structure determination technique. Its most prolific branch is single particle analysis (SPA), a method being used in a growing number of ...Cryo-electron microscopy (cryo-EM) has emerged as a powerful structure determination technique. Its most prolific branch is single particle analysis (SPA), a method being used in a growing number of laboratories worldwide to determine high-resolution protein structures. Cryo-electron tomography (cryo-ET) is another powerful approach that enables visualization of protein complexes in their native cellular environment. Despite the wide-ranging success of cryo-EM, there are many methodological aspects that could be improved. Those include sample preparation, sample screening, data acquisition, image processing, and structure validation. Future developments will increase the reliability and throughput of the technique and reduce the cost and skill level barrier for its adoption. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9599.map.gz emd_9599.map.gz | 166.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9599-v30.xml emd-9599-v30.xml emd-9599.xml emd-9599.xml | 16.7 KB 16.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9599.png emd_9599.png | 250.1 KB | ||
| Masks |  emd_9599_msk_1.map emd_9599_msk_1.map | 178 MB |  Mask map Mask map | |
| Others |  emd_9599_half_map_1.map.gz emd_9599_half_map_1.map.gz emd_9599_half_map_2.map.gz emd_9599_half_map_2.map.gz | 135.4 MB 135.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9599 http://ftp.pdbj.org/pub/emdb/structures/EMD-9599 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9599 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9599 | HTTPS FTP |
-Validation report
| Summary document |  emd_9599_validation.pdf.gz emd_9599_validation.pdf.gz | 78.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9599_full_validation.pdf.gz emd_9599_full_validation.pdf.gz | 77.2 KB | Display | |
| Data in XML |  emd_9599_validation.xml.gz emd_9599_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9599 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9599 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9599 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9599 | HTTPS FTP |
-Related structure data
| Related structure data |  9914C C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10216 (Title: Cryo-EM structure of mouse heavy-chain apoferritin at 1.62 A EMPIAR-10216 (Title: Cryo-EM structure of mouse heavy-chain apoferritin at 1.62 AData size: 3.0 TB / Data #1: RAW movies [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9599.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9599.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | mouse heavy chain apoferritin sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.517 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
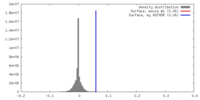
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_9599_msk_1.map emd_9599_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
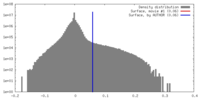
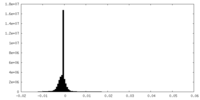
| Density Histograms |
-Half map: mouse heavy chain apoferritin unfiltered half map 2
| File | emd_9599_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | mouse heavy chain apoferritin unfiltered half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
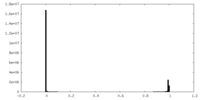
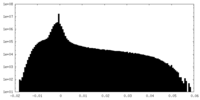
| Density Histograms |
-Half map: mouse heavy chain apoferritin unfiltered half map 1
| File | emd_9599_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | mouse heavy chain apoferritin unfiltered half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
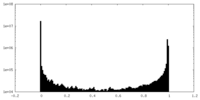
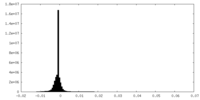
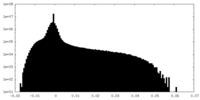
| Density Histograms |
- Sample components
Sample components
-Entire : apoferritin
| Entire | Name: apoferritin |
|---|---|
| Components |
|
-Supramolecule #1: apoferritin
| Supramolecule | Name: apoferritin / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 500 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 4 mg/mL |
|---|---|
| Buffer | pH: 7 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Details: JEOL Ion Bombarder on Soft setting |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 4 K / Instrument: FEI VITROBOT MARK IV / Details: blot for 5 s. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Alignment procedure | Coma free - Residual tilt: 0.02 mrad |
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Sampling interval: 15.0 µm / Number grids imaged: 1 / Number real images: 1647 / Average exposure time: 18.21 sec. / Average electron dose: 50.0 e/Å2 / Details: Exposure rate 0.74 e/pix/s |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 1.3 µm / Calibrated defocus min: 0.2 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.3 µm / Nominal defocus min: 0.2 µm / Nominal magnification: 155000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Refinement | Protocol: RIGID BODY FIT |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)