[English] 日本語
 Yorodumi
Yorodumi- EMDB-22347: CryoEM structure of human light chain apoferritin calculated from... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22347 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of human light chain apoferritin calculated from EER movies (super-resolution experiment, 2x supersampling with random subpixel information) | |||||||||
 Map data Map data | Sharpened map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
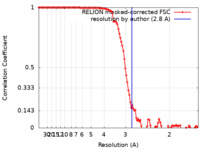
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Guo H / Rubinstein J | |||||||||
| Funding support |  Canada, 2 items Canada, 2 items
| |||||||||
 Citation Citation |  Journal: IUCrJ / Year: 2020 Journal: IUCrJ / Year: 2020Title: Electron-event representation data enable efficient cryoEM file storage with full preservation of spatial and temporal resolution. Authors: Hui Guo / Erik Franken / Yuchen Deng / Samir Benlekbir / Garbi Singla Lezcano / Bart Janssen / Lingbo Yu / Zev A Ripstein / Yong Zi Tan / John L Rubinstein /   Abstract: Direct detector device (DDD) cameras have revolutionized electron cryomicroscopy (cryoEM) with their high detective quantum efficiency (DQE) and output of movie data. A high ratio of camera frame ...Direct detector device (DDD) cameras have revolutionized electron cryomicroscopy (cryoEM) with their high detective quantum efficiency (DQE) and output of movie data. A high ratio of camera frame rate (frames per second) to camera exposure rate (electrons per pixel per second) allows electron counting, which further improves the DQE and enables the recording of super-resolution information. Movie output also allows the correction of specimen movement and compensation for radiation damage. However, these movies come at the cost of producing large volumes of data. It is common practice to sum groups of successive camera frames to reduce the final frame rate, and therefore the file size, to one suitable for storage and image processing. This reduction in the temporal resolution of the camera requires decisions to be made during data acquisition that may result in the loss of information that could have been advantageous during image analysis. Here, experimental analysis of a new electron-event representation (EER) data format for electron-counting DDD movies is presented, which is enabled by new hardware developed by Thermo Fisher Scientific for their Falcon DDD cameras. This format enables the recording of DDD movies at the raw camera frame rate without sacrificing either spatial or temporal resolution. Experimental data demonstrate that the method retains super-resolution information and allows the correction of specimen movement at the physical frame rate of the camera while maintaining manageable file sizes. The EER format will enable the development of new methods that can utilize the full spatial and temporal resolution of DDD cameras. #1:  Journal: Biorxiv / Year: 2020 Journal: Biorxiv / Year: 2020Title: Electron Event Representation (EER) data enables efficient cryoEM file storage with full preservation of spatial and temporal resolution Authors: Guo H / Franken E / Deng Y / Benlekbir S / Lezcano GS / Janssen B / Yu L / Ripstein ZA / Tan YZ / Rubinstein JL | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22347.map.gz emd_22347.map.gz | 116.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22347-v30.xml emd-22347-v30.xml emd-22347.xml emd-22347.xml | 18.8 KB 18.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22347_fsc.xml emd_22347_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_22347.png emd_22347.png | 125.3 KB | ||
| Masks |  emd_22347_msk_1.map emd_22347_msk_1.map | 125 MB |  Mask map Mask map | |
| Others |  emd_22347_half_map_1.map.gz emd_22347_half_map_1.map.gz emd_22347_half_map_2.map.gz emd_22347_half_map_2.map.gz | 115.2 MB 115.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22347 http://ftp.pdbj.org/pub/emdb/structures/EMD-22347 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22347 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22347 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10474 (Title: EER movies of human light chain apoferritin / Data size: 642.1 EMPIAR-10474 (Title: EER movies of human light chain apoferritin / Data size: 642.1 Data #1: Falcon3 EER movies of human light chain apoferritin at 75000 nominal magnification used for intra-fraction motion correction experiment [micrographs - multiframe] Data #2: Falcon4 EER movies of human light chain apoferritin at 47000 nominal magnification used for super-resolution experiment [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_22347.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22347.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_22347_msk_1.map emd_22347_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
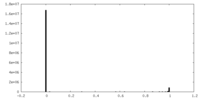
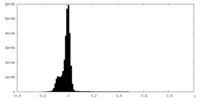
| Density Histograms |
-Half map: Half map 1
| File | emd_22347_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
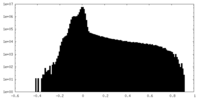
| Density Histograms |
-Half map: Half map 2
| File | emd_22347_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
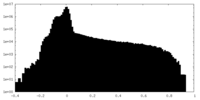
| Density Histograms |
- Sample components
Sample components
-Entire : Human light chain apoferritin
| Entire | Name: Human light chain apoferritin |
|---|---|
| Components |
|
-Supramolecule #1: Human light chain apoferritin
| Supramolecule | Name: Human light chain apoferritin / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293F Homo sapiens (human) / Recombinant cell: HEK293F |
| Molecular weight | Theoretical: 480 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Homemade / Material: COPPER/RHODIUM / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 30.0 nm / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 295 K / Instrument: GATAN CRYOPLUNGE 3 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 100 / Average exposure time: 24.0 sec. / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 1.1 µm / Calibrated defocus min: 0.6 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 47000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)