[English] 日本語
 Yorodumi
Yorodumi- EMDB-32299: Cryo-EM structure of the gastric proton pump complexed with revaprazan -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-32299 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the gastric proton pump complexed with revaprazan | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cation pump / P-type ATPase / gastric / proton pump / membrane protein / P-CAB / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationH+/K+-exchanging ATPase / potassium:proton exchanging ATPase complex / P-type potassium:proton transporter activity / Ion transport by P-type ATPases / P-type sodium:potassium-exchanging transporter activity / sodium:potassium-exchanging ATPase complex / sodium ion export across plasma membrane / intracellular sodium ion homeostasis / potassium ion import across plasma membrane / intracellular potassium ion homeostasis ...H+/K+-exchanging ATPase / potassium:proton exchanging ATPase complex / P-type potassium:proton transporter activity / Ion transport by P-type ATPases / P-type sodium:potassium-exchanging transporter activity / sodium:potassium-exchanging ATPase complex / sodium ion export across plasma membrane / intracellular sodium ion homeostasis / potassium ion import across plasma membrane / intracellular potassium ion homeostasis / potassium ion binding / ATPase activator activity / potassium ion transmembrane transport / proton transmembrane transport / cell adhesion / apical plasma membrane / magnesium ion binding / ATP hydrolysis activity / ATP binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
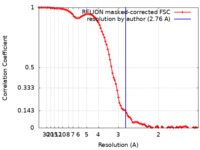
| Method | single particle reconstruction / cryo EM / Resolution: 2.76 Å | |||||||||
 Authors Authors | Abe K / Tanaka S | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: J Med Chem / Year: 2022 Journal: J Med Chem / Year: 2022Title: Structural Basis for Binding of Potassium-Competitive Acid Blockers to the Gastric Proton Pump. Authors: Saki Tanaka / Mikio Morita / Tatsuya Yamagishi / Hridya Valia Madapally / Kenichi Hayashida / Himanshu Khandelia / Christoph Gerle / Hideki Shigematsu / Atsunori Oshima / Kazuhiro Abe /   Abstract: As specific inhibitors of the gastric proton pump, responsible for gastric acidification, K-competitive acid blockers (P-CABs) have recently been utilized in the clinical treatment of gastric acid- ...As specific inhibitors of the gastric proton pump, responsible for gastric acidification, K-competitive acid blockers (P-CABs) have recently been utilized in the clinical treatment of gastric acid-related diseases in Asia. However, as these compounds have been developed based on phenotypic screening, their detailed binding poses are unknown. We show crystal and cryo-EM structures of the gastric proton pump in complex with four different P-CABs, tegoprazan, soraprazan, PF-03716556 and revaprazan, at resolutions reaching 2.8 Å. The structures describe molecular details of their interactions and are supported by functional analyses of mutations and molecular dynamics simulations. We reveal that revaprazan has a novel binding mode in which its tetrahydroisoquinoline moiety binds deep in the cation transport conduit. The mechanism of action of these P-CABs can now be evaluated at the molecular level, which will facilitate the rational development and improvement of currently available P-CABs to provide better treatment of acid-related gastrointestinal diseases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32299.map.gz emd_32299.map.gz | 9.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32299-v30.xml emd-32299-v30.xml emd-32299.xml emd-32299.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_32299_fsc.xml emd_32299_fsc.xml | 9.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_32299.png emd_32299.png | 81.4 KB | ||
| Masks |  emd_32299_msk_1.map emd_32299_msk_1.map | 76.8 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-32299.cif.gz emd-32299.cif.gz | 6.6 KB | ||
| Others |  emd_32299_half_map_1.map.gz emd_32299_half_map_1.map.gz emd_32299_half_map_2.map.gz emd_32299_half_map_2.map.gz | 59.8 MB 60 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32299 http://ftp.pdbj.org/pub/emdb/structures/EMD-32299 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32299 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32299 | HTTPS FTP |
-Related structure data
| Related structure data |  7w4aMC  7w47C  7w48C  7w49C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-11057 (Title: Cryo-EM structure of the gastric proton pump complexed with revaprazan EMPIAR-11057 (Title: Cryo-EM structure of the gastric proton pump complexed with revaprazanData size: 2.1 TB Data #1: K3 movies for the gastric proton pump complexed with revaprazan - Movies_5 [micrographs - multiframe] Data #2: K3 movies for the gastric proton pump complexed with revaprazan - Movies_7 [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_32299.map.gz / Format: CCP4 / Size: 76.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32299.map.gz / Format: CCP4 / Size: 76.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.752 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_32299_msk_1.map emd_32299_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_32299_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_32299_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : gastric proton pump
| Entire | Name: gastric proton pump |
|---|---|
| Components |
|
-Supramolecule #1: gastric proton pump
| Supramolecule | Name: gastric proton pump / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 135 KDa |
-Macromolecule #1: Potassium-transporting ATPase alpha chain 1
| Macromolecule | Name: Potassium-transporting ATPase alpha chain 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: H+/K+-exchanging ATPase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 114.456734 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGKAENYELY QVELGPGPSG DMAAKMSKKK AGRGGGKRKE KLENMKKEME INDHQLSVAE LEQKYQTSAT KGLSASLAAE LLLRDGPNA LRPPRGTPEY VKFARQLAGG LQCLMWVAAA ICLIAFAIQA SEGDLTTDDN LYLALALIAV VVVTGCFGYY Q EFKSTNII ...String: MGKAENYELY QVELGPGPSG DMAAKMSKKK AGRGGGKRKE KLENMKKEME INDHQLSVAE LEQKYQTSAT KGLSASLAAE LLLRDGPNA LRPPRGTPEY VKFARQLAGG LQCLMWVAAA ICLIAFAIQA SEGDLTTDDN LYLALALIAV VVVTGCFGYY Q EFKSTNII ASFKNLVPQQ ATVIRDGDKF QINADQLVVG DLVEMKGGDR VPADIRILQA QGCKVDNSSL TGESEPQTRS PE CTHESPL ETRNIAFFST MCLEGTAQGL VVNTGDRTII GRIASLASGV ENEKTPIAIE IEHFVDIIAG LAILFGATFF IVA MCIGYT FLRAMVFFMA IVVAYVPEGL LATVTVCLSL TAKRLASKNC VVKNLEAVET LGSTSVICS(BFD) KTGTLTQNRM TVSHLWFDN HIHSADTTED QSGQTFDQSS ETWRALCRVL TLCNRAAFKS GQDAVPVPKR IVIGDASETA LLKFSELTLG N AMGYRERF PKVCEIPFNS TNKFQLSIHT LEDPRDPRHV LVMKGAPERV LERCSSILIK GQELPLDEQW REAFQTAYLS LG GLGERVL GFCQLYLSEK DYPPGYAFDV EAMNFPTSGL CFAGLVSMID PPRATVPDAV LKCRTAGIRV IMVTGDHPIT AKA IAASVG IISEGSETVE DIAARLRVPV DQVNRKDARA CVINGMQLKD MDPSELVEAL RTHPEMVFAR TSPQQKLVIV ESCQ RLGAI VAVTGDGVND SPALKKADIG VAMGIAGSDA AKNAADMILL DDNFASIVTG VEQGRLIFDN LKKSIAYTLT KNIPE LTPY LIYITVSVPL PLGCITILFI ELCTDIFPSV SLAYEKAESD IMHLRPRNPK RDRLVNEPLA AYSYFQIGAI QSFAGF TDY FTAMAQEGWF PLLCVGLRPQ WENHHLQDLQ DSYGQEWTFG QRLYQQYTCY TVFFISIEMC QIADVLIRKT RRLSAFQ QG FFRNRILVIA IVFQVCIGCF LCYCPGMPNI FNFMPIRFQW WLVPMPFSLL IFVYDEIRKL GVRCCPGSWW DQELYY UniProtKB: Potassium-transporting ATPase alpha chain 1 |
-Macromolecule #2: Potassium-transporting ATPase subunit beta
| Macromolecule | Name: Potassium-transporting ATPase subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 33.113844 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MAALQEKKSC SQRMEEFQRY CWNPDTGQML GRTLSRWVWI SLYYVAFYVV MSGIFALCIY VLMRTIDPYT PDYQDQLKSP GVTLRPDVY GEKGLDISYN VSDSTTWAGL AHTLHRFLAG YSPAAQEGSI NCTSEKYFFQ ESFLAPNHTK FSCKFTADML Q NCSGRPDP ...String: MAALQEKKSC SQRMEEFQRY CWNPDTGQML GRTLSRWVWI SLYYVAFYVV MSGIFALCIY VLMRTIDPYT PDYQDQLKSP GVTLRPDVY GEKGLDISYN VSDSTTWAGL AHTLHRFLAG YSPAAQEGSI NCTSEKYFFQ ESFLAPNHTK FSCKFTADML Q NCSGRPDP TFGFAEGKPC FIIKMNRIVK FLPGNSTAPR VDCAFLDQPR DGPPLQVEYF PANGTYSLHY FPYYGKKAQP HY SNPLVAA KLLNVPRNRD VVIVCKILAE HVSFDNPHDP YEGKVEFKLK IQK UniProtKB: Potassium-transporting ATPase subunit beta |
-Macromolecule #3: N-(4-fluorophenyl)-4,5-dimethyl-6-[(1R)-1-methyl-3,4-dihydro-1H-i...
| Macromolecule | Name: N-(4-fluorophenyl)-4,5-dimethyl-6-[(1R)-1-methyl-3,4-dihydro-1H-isoquinolin-2-yl]pyrimidin-2-amine type: ligand / ID: 3 / Number of copies: 1 / Formula: 8CK |
|---|---|
| Molecular weight | Theoretical: 362.443 Da |
| Chemical component information | 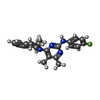 ChemComp-8CK: |
-Macromolecule #4: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 4 / Number of copies: 1 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 6 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #7: 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE
| Macromolecule | Name: 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE / type: ligand / ID: 7 / Number of copies: 1 / Formula: PCW |
|---|---|
| Molecular weight | Theoretical: 787.121 Da |
| Chemical component information |  ChemComp-PCW: |
-Macromolecule #8: water
| Macromolecule | Name: water / type: ligand / ID: 8 / Number of copies: 12 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)