登録情報 データベース : EMDB / ID : EMD-9787タイトル Structure of pancreatic ATP-sensitive potassium channel bound with repaglinide and ATPgammaS at 3.3A resolution the sharpened map from cisTEM 複合体 : KATPタンパク質・ペプチド : ATP-sensitive inward rectifier potassium channel 11タンパク質・ペプチド : ATP-binding cassette sub-family C member 8 isoform X2リガンド : (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphateリガンド : PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTERリガンド : Digitoninリガンド : Repaglinideリガンド : PHOSPHATIDYLETHANOLAMINE / / / / / / / / 機能・相同性 分子機能 ドメイン・相同性 構成要素
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / 生物種 Mus musculus (ハツカネズミ) / Mesocricetus auratus (ネズミ)手法 / / 解像度 : 3.3 Å Chen L / Ding D 資金援助 Organization Grant number 国 Ministry of Science and Technology (China) 2016YFA0502004 National Natural Science Foundation of China 31622021 National Natural Science Foundation of China 31821091 National Natural Science Foundation of China 31870833
ジャーナル : Cell Rep / 年 : 2019タイトル : The Structural Basis for the Binding of Repaglinide to the Pancreatic K Channel.著者 : Dian Ding / Mengmeng Wang / Jing-Xiang Wu / Yunlu Kang / Lei Chen / 要旨 : Repaglinide (RPG) is a short-acting insulin secretagogue widely prescribed for the treatment of type 2 diabetes. It boosts insulin secretion by inhibiting the pancreatic ATP-sensitive potassium ... Repaglinide (RPG) is a short-acting insulin secretagogue widely prescribed for the treatment of type 2 diabetes. It boosts insulin secretion by inhibiting the pancreatic ATP-sensitive potassium channel (K). However, the mechanisms by which RPG binds to the K channel are poorly understood. Here, we describe two cryo-EM structures: the pancreatic K channel in complex with inhibitory RPG and adenosine-5'-(γ-thio)-triphosphate (ATPγS) at 3.3 Å and a medium-resolution structure of a RPG-bound mini SUR1 protein in which the N terminus of the inward-rectifying potassium channel 6.1 (Kir6.1) is fused to the ABC transporter module of the sulfonylurea receptor 1 (SUR1). These structures reveal the binding site of RPG in the SUR1 subunit. Furthermore, the high-resolution structure reveals the complex architecture of the ATP binding site, which is formed by both Kir6.2 and SUR1 subunits, and the domain-domain interaction interfaces. 履歴 登録 2019年1月25日 - ヘッダ(付随情報) 公開 2019年5月22日 - マップ公開 2019年5月22日 - 更新 2024年11月13日 - 現状 2024年11月13日 処理サイト : PDBj / 状態 : 公開
すべて表示 表示を減らす
 データを開く
データを開く 基本情報
基本情報 マップデータ
マップデータ 試料
試料 キーワード
キーワード 機能・相同性情報
機能・相同性情報
 Mesocricetus auratus (ネズミ)
Mesocricetus auratus (ネズミ) データ登録者
データ登録者 中国, 4件
中国, 4件  引用
引用 ジャーナル: Cell Rep / 年: 2019
ジャーナル: Cell Rep / 年: 2019
 構造の表示
構造の表示 ムービービューア
ムービービューア SurfView
SurfView Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク emd_9787.map.gz
emd_9787.map.gz EMDBマップデータ形式
EMDBマップデータ形式 emd-9787-v30.xml
emd-9787-v30.xml emd-9787.xml
emd-9787.xml EMDBヘッダ
EMDBヘッダ emd_9787.png
emd_9787.png emd-9787.cif.gz
emd-9787.cif.gz emd_9787_additional.map.gz
emd_9787_additional.map.gz http://ftp.pdbj.org/pub/emdb/structures/EMD-9787
http://ftp.pdbj.org/pub/emdb/structures/EMD-9787 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9787
ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9787 emd_9787_validation.pdf.gz
emd_9787_validation.pdf.gz EMDB検証レポート
EMDB検証レポート emd_9787_full_validation.pdf.gz
emd_9787_full_validation.pdf.gz emd_9787_validation.xml.gz
emd_9787_validation.xml.gz emd_9787_validation.cif.gz
emd_9787_validation.cif.gz https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9787
https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9787 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9787
ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9787 リンク
リンク EMDB (EBI/PDBe) /
EMDB (EBI/PDBe) /  EMDataResource
EMDataResource マップ
マップ ダウンロード / ファイル: emd_9787.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES)
ダウンロード / ファイル: emd_9787.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) 試料の構成要素
試料の構成要素

 Homo sapiens (ヒト)
Homo sapiens (ヒト) Mesocricetus auratus (ネズミ)
Mesocricetus auratus (ネズミ) Homo sapiens (ヒト)
Homo sapiens (ヒト)


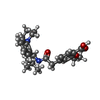

 解析
解析 試料調製
試料調製 電子顕微鏡法
電子顕微鏡法 FIELD EMISSION GUN
FIELD EMISSION GUN
 画像解析
画像解析 ムービー
ムービー コントローラー
コントローラー























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






























