[English] 日本語
 Yorodumi
Yorodumi- EMDB-6832: Structure of pancreatic ATP-sensitive potassium channel bound wit... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6832 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of pancreatic ATP-sensitive potassium channel bound with glibenclamide and ATPgammaS (3D class1 at 4.33A) | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | KATP / channel / glibenclamide / sulfonylurea / MEMBRANE PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationATP sensitive Potassium channels / ATP-activated inward rectifier potassium channel activity / glutamate secretion, neurotransmission / response to resveratrol / inward rectifying potassium channel / Regulation of insulin secretion / sulfonylurea receptor activity / ventricular cardiac muscle tissue development / cell body fiber / ABC-family proteins mediated transport ...ATP sensitive Potassium channels / ATP-activated inward rectifier potassium channel activity / glutamate secretion, neurotransmission / response to resveratrol / inward rectifying potassium channel / Regulation of insulin secretion / sulfonylurea receptor activity / ventricular cardiac muscle tissue development / cell body fiber / ABC-family proteins mediated transport / CAMKK-AMPK signaling cascade / voltage-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / ATPase-coupled monoatomic cation transmembrane transporter activity / inward rectifier potassium channel activity / Ion homeostasis / nervous system process / : / ankyrin binding / neuromuscular process / response to testosterone / response to ATP / potassium ion import across plasma membrane / potassium ion binding / response to stress / action potential / intercalated disc / axolemma / potassium channel activity / ABC-type transporter activity / positive regulation of insulin secretion involved in cellular response to glucose stimulus / heat shock protein binding / cellular response to nutrient levels / T-tubule / acrosomal vesicle / response to ischemia / determination of adult lifespan / positive regulation of protein localization to plasma membrane / cellular response to glucose stimulus / negative regulation of insulin secretion / ADP binding / sarcolemma / cellular response to nicotine / glucose metabolic process / cellular response to tumor necrosis factor / response to estradiol / nuclear envelope / presynapse / presynaptic membrane / transmembrane transporter binding / response to hypoxia / endosome / response to xenobiotic stimulus / neuronal cell body / apoptotic process / glutamatergic synapse / ATP hydrolysis activity / ATP binding / metal ion binding / plasma membrane / cytoplasm Similarity search - Function | ||||||||||||||||||
| Biological species |   Mesocricetus auratus (golden hamster) Mesocricetus auratus (golden hamster) | ||||||||||||||||||
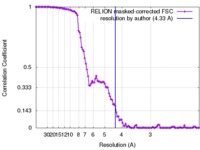
| Method | single particle reconstruction / cryo EM / Resolution: 4.33 Å | ||||||||||||||||||
 Authors Authors | Chen L / Wu JX | ||||||||||||||||||
| Funding support |  China, 5 items China, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Protein Cell / Year: 2018 Journal: Protein Cell / Year: 2018Title: Ligand binding and conformational changes of SUR1 subunit in pancreatic ATP-sensitive potassium channels. Authors: Jing-Xiang Wu / Dian Ding / Mengmeng Wang / Yunlu Kang / Xin Zeng / Lei Chen /  Abstract: ATP-sensitive potassium channels (K) are energy sensors on the plasma membrane. By sensing the intracellular ADP/ATP ratio of β-cells, pancreatic K channels control insulin release and regulate ...ATP-sensitive potassium channels (K) are energy sensors on the plasma membrane. By sensing the intracellular ADP/ATP ratio of β-cells, pancreatic K channels control insulin release and regulate metabolism at the whole body level. They are implicated in many metabolic disorders and diseases and are therefore important drug targets. Here, we present three structures of pancreatic K channels solved by cryo-electron microscopy (cryo-EM), at resolutions ranging from 4.1 to 4.5 Å. These structures depict the binding site of the antidiabetic drug glibenclamide, indicate how Kir6.2 (inward-rectifying potassium channel 6.2) N-terminus participates in the coupling between the peripheral SUR1 (sulfonylurea receptor 1) subunit and the central Kir6.2 channel, reveal the binding mode of activating nucleotides, and suggest the mechanism of how Mg-ADP binding on nucleotide binding domains (NBDs) drives a conformational change of the SUR1 subunit. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6832.map.gz emd_6832.map.gz | 12.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6832-v30.xml emd-6832-v30.xml emd-6832.xml emd-6832.xml | 17.9 KB 17.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_6832_fsc.xml emd_6832_fsc.xml | 10.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_6832.png emd_6832.png | 185.7 KB | ||
| Filedesc metadata |  emd-6832.cif.gz emd-6832.cif.gz | 6.6 KB | ||
| Others |  emd_6832_half_map_1.map.gz emd_6832_half_map_1.map.gz emd_6832_half_map_2.map.gz emd_6832_half_map_2.map.gz | 86.7 MB 86.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6832 http://ftp.pdbj.org/pub/emdb/structures/EMD-6832 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6832 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6832 | HTTPS FTP |
-Related structure data
| Related structure data |  5ykfMC  6831C  6833C  6847C  6848C  6849C  6850C  6851C  6852C  6853C  5ykeC  5ykgC  5yw7C  5yw8C  5yw9C  5ywaC  5ywbC  5ywcC  5ywdC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6832.map.gz / Format: CCP4 / Size: 115.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6832.map.gz / Format: CCP4 / Size: 115.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.055 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
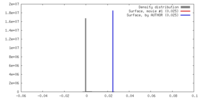
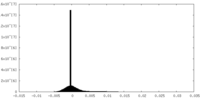
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #1
| File | emd_6832_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
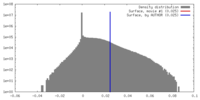
| Density Histograms |
-Half map: #2
| File | emd_6832_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
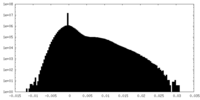
| Density Histograms |
- Sample components
Sample components
-Entire : KATP
| Entire | Name: KATP |
|---|---|
| Components |
|
-Supramolecule #1: KATP
| Supramolecule | Name: KATP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: ATP-sensitive inward rectifier potassium channel 11
| Macromolecule | Name: ATP-sensitive inward rectifier potassium channel 11 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 43.615734 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MLSRKGIIPE EYVLTRLAED PAEPRYRTRE RRARFVSKKG NCNVAHKNIR EQGRFLQDVF TTLVDLKWPH TLLIFTMSFL CSWLLFAMV WWLIAFAHGD LAPGEGTNVP CVTSIHSFSS AFLFSIEVQV TIGFGGRMVT EECPLAILIL IVQNIVGLMI N AIMLGCIF ...String: MLSRKGIIPE EYVLTRLAED PAEPRYRTRE RRARFVSKKG NCNVAHKNIR EQGRFLQDVF TTLVDLKWPH TLLIFTMSFL CSWLLFAMV WWLIAFAHGD LAPGEGTNVP CVTSIHSFSS AFLFSIEVQV TIGFGGRMVT EECPLAILIL IVQNIVGLMI N AIMLGCIF MKTAQAHRRA ETLIFSKHAV ITLRHGRLCF MLRVGDLRKS MIISATIHMQ VVRKTTSPEG EVVPLHQVDI PM ENGVGGN GIFLVAPLII YHVIDSNSPL YDLAPSDLHH HQDLEIIVIL EGVVETTGIT TQARTSYLAD EILWGQRFVP IVA EEDGRY SVDYSKFGNT IKVPTPLCTA RQLDEDRSLL DALTLASSRG PLRKRSVAVA KAKPKFSISP DSLS UniProtKB: ATP-sensitive inward rectifier potassium channel 11 |
-Macromolecule #2: ATP-binding cassette sub-family C member 8 isoform X2
| Macromolecule | Name: ATP-binding cassette sub-family C member 8 isoform X2 / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mesocricetus auratus (golden hamster) Mesocricetus auratus (golden hamster) |
| Molecular weight | Theoretical: 177.295516 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MPLAFCGTEN HSAAYRVDQG VLNNGCFVDA LNVVPHVFLL FITFPILFIG WGSQSSKVHI HHSTWLHFPG HNLRWILTFI LLFVLVCEI AEGILSDGVT ESRHLHLYMP AGMAFMAAIT SVVYYHNIET SNFPKLLIAL LIYWTLAFIT KTIKFVKFYD H AIGFSQLR ...String: MPLAFCGTEN HSAAYRVDQG VLNNGCFVDA LNVVPHVFLL FITFPILFIG WGSQSSKVHI HHSTWLHFPG HNLRWILTFI LLFVLVCEI AEGILSDGVT ESRHLHLYMP AGMAFMAAIT SVVYYHNIET SNFPKLLIAL LIYWTLAFIT KTIKFVKFYD H AIGFSQLR FCLTGLLVIL YGMLLLVEVN VIRVRRYIFF KTPREVKPPE DLQDLGVRFL QPFVNLLSKG TYWWMNAFIK TA HKKPIDL RAIGKLPIAM RALTNYQRLC VAFDAQARKD TQSPQGARAI WRALCHAFGR RLILSSTFRI LADLLGFAGP LCI FGIVDH LGKENHVFQP KTQFLGVYFV SSQEFLGNAY VLAVLLFLAL LLQRTFLQAS YYVAIETGIN LRGAIQTKIY NKIM HLSTS NLSMGEMTAG QICNLVAIDT NQLMWFFFLC PNLWAMPVQI IVGVILLYYI LGVSALIGAA VIILLAPVQY FVATK LSQA QRSTLEHSNE RLKQTNEMLR GMKLLKLYAW ESIFCSRVEV TRRKEMTSLR AFAVYTSISI FMNTAIPIAA VLITFV GHV SFFKESDLSP SVAFASLSLF HILVTPLFLL SSVVRSTVKA LVSVQKLSEF LSSAEIREEQ CAPREPAPQG QAGKYQA VP LKVVNRKRPA REEVRDLLGP LQRLAPSMDG DADNFCVQII GGFFTWTPDG IPTLSNITIR IPRGQLTMIV GQVGCGKS S LLLATLGEMQ KVSGAVFWNS NLPDSEGEDP SSPERETAAG SDIRSRGPVA YASQKPWLLN ATVEENITFE SPFNKQRYK MVIEACSLQP DIDILPHGDQ TQIGERGINL SGGQRQRISV ARALYQQTNV VFLDDPFSAL DVHLSDHLMQ AGILELLRDD KRTVVLVTH KLQYLPHADW IIAMKDGTIQ REGTLKDFQR SECQLFEHWK TLMNRQDQEL EKETVMERKA SEPSQGLPRA M SSRDGLLL DEEEEEEEAA ESEEDDNLSS VLHQRAKIPW RACTKYLSSA GILLLSLLVF SQLLKHMVLV AIDYWLAKWT DS ALVLSPA ARNCSLSQEC DLDQSVYAMV FTLLCSLGIV LCLVTSVTVE WTGLKVAKRL HRSLLNRIIL APMRFFETTP LGS ILNRFS SDCNTIDQHI PSTLECLSRS TLLCVSALTV ISYVTPVFLV ALLPLAVVCY FIQKYFRVAS RDLQQLDDTT QLPL LSHFA ETVEGLTTIR AFRYEARFQQ KLLEYTDSNN IASLFLTAAN RWLEVRMEYI GACVVLIAAA TSISNSLHRE LSAGL VGLG LTYALMVSNY LNWMVRNLAD MEIQLGAVKR IHALLKTEAE SYEGLLAPSL IPKNWPDQGK IQIQNLSVRY DSSLKP VLK HVNALISPGQ KIGICGRTGS GKSSFSLAFF RMVDMFEGRI IIDGIDIAKL PLHTLRSRLS IILQDPVLFS GTIRFNL DP EKKCSDSTLW EALEIAQLKL VVKALPGGLD AIITEGGENF SQGQRQLFCL ARAFVRKTSI FIMDEATASI DMATENIL Q KVVMTAFADR TVVTIAHRVH TILSADLVMV LKRGAILEFD KPETLLSQKD SVFASFVRAD K UniProtKB: ATP-binding cassette sub-family C member 8 |
-Macromolecule #3: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER / type: ligand / ID: 3 / Number of copies: 8 / Formula: AGS |
|---|---|
| Molecular weight | Theoretical: 523.247 Da |
| Chemical component information |  ChemComp-AGS: |
-Macromolecule #4: 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-...
| Macromolecule | Name: 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide type: ligand / ID: 4 / Number of copies: 4 / Formula: GBM |
|---|---|
| Molecular weight | Theoretical: 494.004 Da |
| Chemical component information | 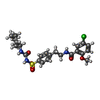 ChemComp-GBM: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)