+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9246 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of full-length IP3R1 channel in Apo-state | |||||||||||||||||||||
 Map data Map data | cryo-EM density map of IP3R1 in apo-state | |||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||
| Biological species |  | |||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.3 Å | |||||||||||||||||||||
 Authors Authors | Fan G / Baker MR / Wang Z / Seryshev AB / Ludtke SJ / Baker ML / Serysheva II | |||||||||||||||||||||
| Funding support |  United States, 6 items United States, 6 items
| |||||||||||||||||||||
 Citation Citation |  Journal: Cell Res / Year: 2018 Journal: Cell Res / Year: 2018Title: Cryo-EM reveals ligand induced allostery underlying InsPR channel gating. Authors: Guizhen Fan / Mariah R Baker / Zhao Wang / Alexander B Seryshev / Steven J Ludtke / Matthew L Baker / Irina I Serysheva /  Abstract: Inositol-1,4,5-trisphosphate receptors (InsPRs) are cation channels that mobilize Ca from intracellular stores in response to a wide range of cellular stimuli. The paradigm of InsPR activation is the ...Inositol-1,4,5-trisphosphate receptors (InsPRs) are cation channels that mobilize Ca from intracellular stores in response to a wide range of cellular stimuli. The paradigm of InsPR activation is the coupled interplay between binding of InsP and Ca that switches the ion conduction pathway between closed and open states to enable the passage of Ca through the channel. However, the molecular mechanism of how the receptor senses and decodes ligand-binding signals into gating motion remains unknown. Here, we present the electron cryo-microscopy structure of InsPR1 from rat cerebellum determined to 4.1 Å resolution in the presence of activating concentrations of Ca and adenophostin A (AdA), a structural mimetic of InsP and the most potent known agonist of the channel. Comparison with the 3.9 Å-resolution structure of InsPR1 in the Apo-state, also reported herein, reveals the binding arrangement of AdA in the tetrameric channel assembly and striking ligand-induced conformational rearrangements within cytoplasmic domains coupled to the dilation of a hydrophobic constriction at the gate. Together, our results provide critical insights into the mechanistic principles by which ligand-binding allosterically gates InsPR channel. | |||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9246.map.gz emd_9246.map.gz | 25 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9246-v30.xml emd-9246-v30.xml emd-9246.xml emd-9246.xml | 13 KB 13 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9246.png emd_9246.png | 232.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9246 http://ftp.pdbj.org/pub/emdb/structures/EMD-9246 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9246 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9246 | HTTPS FTP |
-Validation report
| Summary document |  emd_9246_validation.pdf.gz emd_9246_validation.pdf.gz | 78.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9246_full_validation.pdf.gz emd_9246_full_validation.pdf.gz | 78 KB | Display | |
| Data in XML |  emd_9246_validation.xml.gz emd_9246_validation.xml.gz | 494 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9246 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9246 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9246 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9246 | HTTPS FTP |
-Related structure data
| Related structure data |  9243C  9244C  9245C  9247C  9248C  6mu1C  6mu2C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9246.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9246.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryo-EM density map of IP3R1 in apo-state | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.26 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
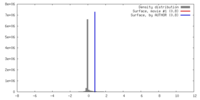
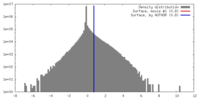
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Inositol 1,4,5-trisphosphate receptor
| Entire | Name: Inositol 1,4,5-trisphosphate receptor |
|---|---|
| Components |
|
-Supramolecule #1: Inositol 1,4,5-trisphosphate receptor
| Supramolecule | Name: Inositol 1,4,5-trisphosphate receptor / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: tetrameric assembly |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1.3 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Details: 50 mM Tris-HCl buffer (pH 7.4), 150 mM NaCl, 1 mM DTT, 0.4% CHAPS, 2 mM EGTA, 1 mM EDTA, protease inhibitors |
|---|---|
| Grid | Support film - Material: CARBON / Support film - topology: CONTINUOUS / Details: unavailable |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
| Details | 0.1 mg/ml |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F30 |
|---|---|
| Temperature | Min: 93.0 K / Max: 93.0 K |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 2-17 / Number real images: 9823 / Average exposure time: 0.2 sec. / Average electron dose: 1.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm |
| Sample stage | Specimen holder model: GATAN 910 MULTI-SPECIMEN SINGLE TILT CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai F30 / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)