[English] 日本語
 Yorodumi
Yorodumi- EMDB-9188: Cryo-EM structure of the human neutral amino acid transporter ASCT2 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9188 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human neutral amino acid transporter ASCT2 | |||||||||
 Map data Map data | human neutral amino acid transporter | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | human neutral amino acid transporter / ASCT2 / outward-oriented state / membrane protein / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationglutamine secretion / L-glutamine import across plasma membrane / L-glutamine transmembrane transporter activity / L-serine transmembrane transporter activity / glutamine transport / ligand-gated channel activity / neutral amino acid transport / L-aspartate transmembrane transporter activity / L-aspartate import across plasma membrane / neutral L-amino acid transmembrane transporter activity ...glutamine secretion / L-glutamine import across plasma membrane / L-glutamine transmembrane transporter activity / L-serine transmembrane transporter activity / glutamine transport / ligand-gated channel activity / neutral amino acid transport / L-aspartate transmembrane transporter activity / L-aspartate import across plasma membrane / neutral L-amino acid transmembrane transporter activity / Amino acid transport across the plasma membrane / symporter activity / amino acid transmembrane transporter activity / antiporter activity / RHOJ GTPase cycle / protein homotrimerization / RHOQ GTPase cycle / amino acid transport / RHOH GTPase cycle / RAC3 GTPase cycle / transport across blood-brain barrier / RAC1 GTPase cycle / basal plasma membrane / erythrocyte differentiation / centriolar satellite / melanosome / signaling receptor activity / virus receptor activity / ciliary basal body / extracellular exosome / metal ion binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.84 Å | |||||||||
 Authors Authors | Yu X / Han S | |||||||||
 Citation Citation |  Journal: Elife / Year: 2019 Journal: Elife / Year: 2019Title: Cryo-EM structures of the human glutamine transporter SLC1A5 (ASCT2) in the outward-facing conformation. Authors: Xiaodi Yu / Olga Plotnikova / Paul D Bonin / Timothy A Subashi / Thomas J McLellan / Darren Dumlao / Ye Che / Yin Yao Dong / Elisabeth P Carpenter / Graham M West / Xiayang Qiu / Jeffrey S Culp / Seungil Han /   Abstract: Alanine-serine-cysteine transporter 2 (ASCT2, SLC1A5) is the primary transporter of glutamine in cancer cells and regulates the mTORC1 signaling pathway. The SLC1A5 function involves finely tuned ...Alanine-serine-cysteine transporter 2 (ASCT2, SLC1A5) is the primary transporter of glutamine in cancer cells and regulates the mTORC1 signaling pathway. The SLC1A5 function involves finely tuned orchestration of two domain movements that include the substrate-binding transport domain and the scaffold domain. Here, we present cryo-EM structures of human SLC1A5 and its complex with the substrate, L-glutamine in an outward-facing conformation. These structures reveal insights into the conformation of the critical ECL2a loop which connects the two domains, thus allowing rigid body movement of the transport domain throughout the transport cycle. Furthermore, the structures provide new insights into substrate recognition, which involves conformational changes in the HP2 loop. A putative cholesterol binding site was observed near the domain interface in the outward-facing state. Comparison with the previously determined inward-facing structure of SCL1A5 provides a basis for a more integrated understanding of substrate recognition and transport mechanism in the SLC1 family. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9188.map.gz emd_9188.map.gz | 3.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9188-v30.xml emd-9188-v30.xml emd-9188.xml emd-9188.xml | 16.1 KB 16.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9188.png emd_9188.png | 59.9 KB | ||
| Masks |  emd_9188_msk_1.map emd_9188_msk_1.map | 42.9 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-9188.cif.gz emd-9188.cif.gz | 5.9 KB | ||
| Others |  emd_9188_half_map_1.map.gz emd_9188_half_map_1.map.gz emd_9188_half_map_2.map.gz emd_9188_half_map_2.map.gz | 2.6 MB 2.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9188 http://ftp.pdbj.org/pub/emdb/structures/EMD-9188 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9188 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9188 | HTTPS FTP |
-Related structure data
| Related structure data |  6mpbMC  9187C  6mp6C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_9188.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9188.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | human neutral amino acid transporter | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
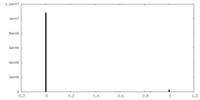
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.086 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
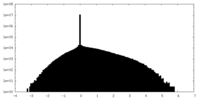
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_9188_msk_1.map emd_9188_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
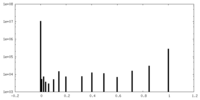
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: human neutral amino acid transporter, half map #1
| File | emd_9188_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | human neutral amino acid transporter, half map #1 | ||||||||||||
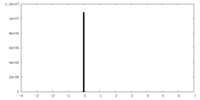
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: human neutral amino acid transporter, half map #2
| File | emd_9188_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | human neutral amino acid transporter, half map #2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ternary complex of SLC1A5
| Entire | Name: Ternary complex of SLC1A5 |
|---|---|
| Components |
|
-Supramolecule #1: Ternary complex of SLC1A5
| Supramolecule | Name: Ternary complex of SLC1A5 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 170 KDa |
-Macromolecule #1: Neutral amino acid transporter B(0)
| Macromolecule | Name: Neutral amino acid transporter B(0) / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 56.638902 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MVADPPRDSK GLAAAEPTAN GGLALASIED QGAAAGGYCG SRDQVRRCLR ANLLVLLTVV AVVAGVALGL GVSGAGGALA LGPERLSAF VFPGELLLRL LRMIILPLVV CSLIGGAASL DPGALGRLGA WALLFFLVTT LLASALGVGL ALALQPGAAS A AINASVGA ...String: MVADPPRDSK GLAAAEPTAN GGLALASIED QGAAAGGYCG SRDQVRRCLR ANLLVLLTVV AVVAGVALGL GVSGAGGALA LGPERLSAF VFPGELLLRL LRMIILPLVV CSLIGGAASL DPGALGRLGA WALLFFLVTT LLASALGVGL ALALQPGAAS A AINASVGA AGSAENAPSK EVLDSFLDLA RNIFPSNLVS AAFRSYSTTY EERNITGTRV KVPVGQEVEG MNILGLVVFA IV FGVALRK LGPEGELLIR FFNSFNEATM VLVSWIMWYA PVGIMFLVAG KIVEMEDVGL LFARLGKYIL CCLLGHAIHG LLV LPLIYF LFTRKNPYRF LWGIVTPLAT AFGTSSSSAT LPLMMKCVEE NNGVAKHISR FILPIGATVN MDGAALFQCV AAVF IAQLS QQSLDFVKII TILVTATASS VGAAGIPAGG VLTLAIILEA VNLPVDHISL ILAVDWLVDR SCTVLNVEGD ALGAG LLQN YVDRTESRST EPELIQVKSE LPLDPLPVPT EEGNPLLKHY RGPAGDATVA SEKESVM UniProtKB: Neutral amino acid transporter B(0) |
-Macromolecule #2: GLUTAMINE
| Macromolecule | Name: GLUTAMINE / type: ligand / ID: 2 / Number of copies: 3 / Formula: GLN |
|---|---|
| Molecular weight | Theoretical: 146.144 Da |
| Chemical component information |  ChemComp-GLN: |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 3 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 1.06 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.84 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cisTEM / Number images used: 253220 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: RELION |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD / Software - Name: cisTEM |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)