+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4898 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | TssA protein from T6SS of Vibrio cholerae. | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | T6SS / Vibrio / TssA / cap / TRANSPORT PROTEIN | |||||||||
| Function / homology | Type VI secretion system-associated, VCA0119 / Type VI secretion, EvfE, EvfF, ImpA, BimE, VC_A0119, VasJ / ImpA, N-terminal / ImpA, N-terminal, type VI secretion system / Type VI secretion system protein TssA / ImpA N-terminal domain-containing protein Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
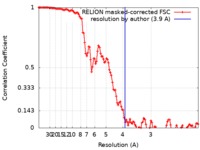
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Nazarov S / Adaixo R / Basler M | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2019 Journal: EMBO J / Year: 2019Title: Diverse roles of TssA-like proteins in the assembly of bacterial type VI secretion systems. Authors: Johannes Paul Schneider / Sergey Nazarov / Ricardo Adaixo / Martina Liuzzo / Peter David Ringel / Henning Stahlberg / Marek Basler /  Abstract: Protein translocation by the bacterial type VI secretion system (T6SS) is driven by a rapid contraction of a sheath assembled around a tube with associated effectors. Here, we show that TssA-like or ...Protein translocation by the bacterial type VI secretion system (T6SS) is driven by a rapid contraction of a sheath assembled around a tube with associated effectors. Here, we show that TssA-like or TagA-like proteins with a conserved N-terminal domain and varying C-terminal domains can be grouped into at least three distinct classes based on their role in sheath assembly. The proteins of the first class increase speed and frequency of sheath assembly and form a stable dodecamer at the distal end of a polymerizing sheath. The proteins of the second class localize to the cell membrane and block sheath polymerization upon extension across the cell. This prevents excessive sheath polymerization and bending, which may result in sheath destabilization and detachment from its membrane anchor and thus result in failed secretion. The third class of these proteins localizes to the baseplate and is required for initiation of sheath assembly. Our work shows that while various proteins share a conserved N-terminal domain, their roles in T6SS biogenesis are fundamentally different. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4898.map.gz emd_4898.map.gz | 100 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4898-v30.xml emd-4898-v30.xml emd-4898.xml emd-4898.xml | 22.6 KB 22.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4898_fsc.xml emd_4898_fsc.xml | 11.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_4898.png emd_4898.png | 179.6 KB | ||
| Masks |  emd_4898_msk_1.map emd_4898_msk_1.map | 129.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-4898.cif.gz emd-4898.cif.gz | 6.3 KB | ||
| Others |  emd_4898_additional_1.map.gz emd_4898_additional_1.map.gz emd_4898_additional_2.map.gz emd_4898_additional_2.map.gz emd_4898_additional_3.map.gz emd_4898_additional_3.map.gz emd_4898_half_map_1.map.gz emd_4898_half_map_1.map.gz emd_4898_half_map_2.map.gz emd_4898_half_map_2.map.gz | 7.5 MB 119.8 MB 120.9 MB 100.4 MB 100.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4898 http://ftp.pdbj.org/pub/emdb/structures/EMD-4898 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4898 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4898 | HTTPS FTP |
-Validation report
| Summary document |  emd_4898_validation.pdf.gz emd_4898_validation.pdf.gz | 676.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_4898_full_validation.pdf.gz emd_4898_full_validation.pdf.gz | 675.9 KB | Display | |
| Data in XML |  emd_4898_validation.xml.gz emd_4898_validation.xml.gz | 18.6 KB | Display | |
| Data in CIF |  emd_4898_validation.cif.gz emd_4898_validation.cif.gz | 24.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4898 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4898 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4898 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4898 | HTTPS FTP |
-Related structure data
| Related structure data |  6riuMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10271 (Title: Cryo electron microscopy of TssA protein from T6SS of Vibrio cholerae. EMPIAR-10271 (Title: Cryo electron microscopy of TssA protein from T6SS of Vibrio cholerae.Data size: 1.3 TB Data #1: Frame-averaged micrographs of TssA protein from T6SS of Vibrio cholerae [micrographs - single frame] Data #2: Aligned multi-frame micrographs of TssA protein from T6SS of Vibrio cholerae [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4898.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4898.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.058 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_4898_msk_1.map emd_4898_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_4898_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #3
| File | emd_4898_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #2
| File | emd_4898_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_4898_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_4898_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Type VI secretion system protein TssA.
| Entire | Name: Type VI secretion system protein TssA. |
|---|---|
| Components |
|
-Supramolecule #1: Type VI secretion system protein TssA.
| Supramolecule | Name: Type VI secretion system protein TssA. / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 634 KDa |
-Supramolecule #2: Middle N-terminal domain (Nt2) of TssA protein from T6SS of Vibri...
| Supramolecule | Name: Middle N-terminal domain (Nt2) of TssA protein from T6SS of Vibrio cholerae. type: complex / ID: 2 / Parent: 1 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Type VI secretion system protein TssA
| Macromolecule | Name: Type VI secretion system protein TssA / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 53.102793 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEMTEYRQCV AKPISSSNPV GERLVDHPLF DFIEDQMMKV GSLSHASVQW DEVEHSTLKL LGEQSKDIKL LVYLLQCLHN QVTPTRLIT SFAVMSEFLN QYWNDSYPAP GARGNLPRRK FFSQMAQRFT TVIEKFDFHH LDEADRQALQ AAVEEWQQAV E KQGLSSEL ...String: MEMTEYRQCV AKPISSSNPV GERLVDHPLF DFIEDQMMKV GSLSHASVQW DEVEHSTLKL LGEQSKDIKL LVYLLQCLHN QVTPTRLIT SFAVMSEFLN QYWNDSYPAP GARGNLPRRK FFSQMAQRFT TVIEKFDFHH LDEADRQALQ AAVEEWQQAV E KQGLSSEL VESVVVRITA EIKRAEQRQQ VTAQSSAERE TPSAATSSPA ASMVVDHSSD KAAKQTLLKV ADFLAEQEFG IA LSIRLRR FAVWGSITSL PDHKPDGETL LRGMQADRVK DYQDQLRHPD LALWRKVEQS LTMAPYWFEG QWMSYTIAQQ LGK SDWCQA IAEETQQFLR RLPSLLELKF KGGEPFVSDS VKEWLASVGQ SQGAAGQSVG GDWQEKRKEA FQLAKEGGIA VALS MLNDG LVSAVEPRDK FYWRLLSADL LRANHLDAMA GEQYQTLLNQ VTTLSVPEWE PSLVEQIQRY TTSE UniProtKB: Type VI secretion system protein TssA |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: LACEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 293 K / Details: Leica EM GP2. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 2963 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)