+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: PDB / ID: 2da8 | ||||||
|---|---|---|---|---|---|---|---|
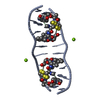
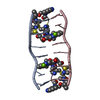
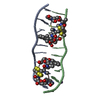
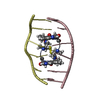
| タイトル | SOLUTION STRUCTURE OF A COMPLEX BETWEEN (N-MECYS3,N-MECYS7)TANDEM AND (D(GATATC))2 | ||||||
 要素 要素 |
| ||||||
 キーワード キーワード | DNA/ANTIBIOTIC / BISINTERCALATOR / DEPSIPEPTIDE / QUINOXALINE / ANTIBIOTIC / ANTITUMOR / DNA-ANTIBIOTIC COMPLEX | ||||||
| 機能・相同性 | 2-CARBOXYQUINOXALINE / : / DNA 機能・相同性情報 機能・相同性情報 | ||||||
| 生物種 |  STREPTOMYCINEAE (バクテリア) STREPTOMYCINEAE (バクテリア) | ||||||
| 手法 | 溶液NMR / ENERGY MINIMIZATION, RESTRAINED MOLECULAR DYNAMICS, NOE BASED RELAXATION REFINEMENT | ||||||
 データ登録者 データ登録者 | Addess, K.J. / Sinsheimer, J.S. / Feigon, J. | ||||||
 引用 引用 |  ジャーナル: Biochemistry / 年: 1993 ジャーナル: Biochemistry / 年: 1993タイトル: Solution Structure of a Complex between [N-Mecys3,N-Mecys7]Tandem and [D(Gatatc)]2. 著者: Addess, K.J. / Sinsheimer, J.S. / Feigon, J. | ||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 構造ビューア | 分子:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- ダウンロードとリンク
ダウンロードとリンク
- ダウンロード
ダウンロード
| PDBx/mmCIF形式 |  2da8.cif.gz 2da8.cif.gz | 88.8 KB | 表示 |  PDBx/mmCIF形式 PDBx/mmCIF形式 |
|---|---|---|---|---|
| PDB形式 |  pdb2da8.ent.gz pdb2da8.ent.gz | 69.5 KB | 表示 |  PDB形式 PDB形式 |
| PDBx/mmJSON形式 |  2da8.json.gz 2da8.json.gz | ツリー表示 |  PDBx/mmJSON形式 PDBx/mmJSON形式 | |
| その他 |  その他のダウンロード その他のダウンロード |
-検証レポート
| アーカイブディレクトリ |  https://data.pdbj.org/pub/pdb/validation_reports/da/2da8 https://data.pdbj.org/pub/pdb/validation_reports/da/2da8 ftp://data.pdbj.org/pub/pdb/validation_reports/da/2da8 ftp://data.pdbj.org/pub/pdb/validation_reports/da/2da8 | HTTPS FTP |
|---|
-関連構造データ
| 関連構造データ | |
|---|---|
| 類似構造データ |
- リンク
リンク
- 集合体
集合体
| 登録構造単位 | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| Atom site foot note | 1: RESIDUES SER A 1 AND SER B 1 ARE D-AMINO ACIDS. 2: ALA A 2 - CYS A 3 MODEL 1 OMEGA = 0.00 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION 3: ALA B 2 - CYS B 3 MODEL 1 OMEGA = 0.00 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION 4: ALA A 2 - CYS A 3 MODEL 2 OMEGA = 0.02 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION 5: ALA A 2 - CYS A 3 MODEL 3 OMEGA = 0.00 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION 6: ALA B 2 - CYS B 3 MODEL 3 OMEGA = 0.00 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION 7: ALA A 2 - CYS A 3 MODEL 4 OMEGA =359.98 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION 8: ALA B 2 - CYS B 3 MODEL 4 OMEGA = 0.00 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION 9: ALA A 2 - CYS A 3 MODEL 5 OMEGA = 0.00 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION 10: ALA B 2 - CYS B 3 MODEL 5 OMEGA = 0.00 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION 11: ALA A 2 - CYS A 3 MODEL 6 OMEGA = 0.02 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION 12: ALA A 2 - CYS A 3 MODEL 7 OMEGA = 0.00 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION 13: ALA A 2 - CYS A 3 MODEL 8 OMEGA = 0.00 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION | |||||||||
| NMR アンサンブル |
|
- 要素
要素
| #1: タンパク質・ペプチド | 分子量: 766.928 Da / 分子数: 1 / 変異: YES / 由来タイプ: 合成 / 詳細: VALINE AT POSITIONS 4 AND 8 / 由来: (合成)  STREPTOMYCINEAE (バクテリア) / 参照: NOR: NOR01129 STREPTOMYCINEAE (バクテリア) / 参照: NOR: NOR01129 | ||
|---|---|---|---|
| #2: DNA鎖 | 分子量: 1808.229 Da / 分子数: 2 / 由来タイプ: 合成 #3: 化合物 | |
-実験情報
-実験
| 実験 | 手法: 溶液NMR |
|---|---|
| NMR実験の詳細 | Text: FOR DISTANCE GEOMETRY, FULL STRUCTURE EMBEDS WERE DONE USING THE PROGRAM DSPACE (HARE RESEARCH, INC.) AND SUBSTRUCTURE EMBEDS WERE DONE USING X-PLOR (A. T. BRUNGER, YALE UNIVERSITY). ALL ...Text: FOR DISTANCE GEOMETRY, FULL STRUCTURE EMBEDS WERE DONE USING THE PROGRAM DSPACE (HARE RESEARCH, INC.) AND SUBSTRUCTURE EMBEDS WERE DONE USING X-PLOR (A. T. BRUNGER, YALE UNIVERSITY). ALL REFINEMENT STEPS WERE DONE USING X-PLOR. |
- 解析
解析
| ソフトウェア |
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NMR software |
| |||||||||
| 精密化 | 手法: ENERGY MINIMIZATION, RESTRAINED MOLECULAR DYNAMICS, NOE BASED RELAXATION REFINEMENT ソフトェア番号: 1 詳細: EIGHT STRUCTURES WERE GENERATED BY DISTANCE GEOMETRY. TWO DIMENSIONAL DERIVED NOE AND DIHEDRAL BOND ANGLE CONSTRAINTS WERE USED THROUGHOUT ALL STAGES OF THE STRUCTURE CALCULATIONS. AVERAGE R ...詳細: EIGHT STRUCTURES WERE GENERATED BY DISTANCE GEOMETRY. TWO DIMENSIONAL DERIVED NOE AND DIHEDRAL BOND ANGLE CONSTRAINTS WERE USED THROUGHOUT ALL STAGES OF THE STRUCTURE CALCULATIONS. AVERAGE R VALUE FOR 8 STRUCTURES 0.21 | |||||||||
| NMRアンサンブル | コンフォーマー選択の基準: all calculated structures submitted 計算したコンフォーマーの数: 8 / 登録したコンフォーマーの数: 8 | |||||||||
| NMR ensemble rms | Atom type: all heavy atoms / Distance rms dev: 1.1 Å |
 ムービー
ムービー コントローラー
コントローラー












 PDBj
PDBj

 X-PLOR
X-PLOR