[English] 日本語
 Yorodumi
Yorodumi- EMDB-24720: SaPIbov5 procapsid structure including size redirecting protein Ccm -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24720 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SaPIbov5 procapsid structure including size redirecting protein Ccm | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HK97-like fold / capsid size redirection / major capsid protein / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationATP-dependent peptidase activity / membrane => GO:0016020 / serine-type endopeptidase activity Similarity search - Function | |||||||||
| Biological species |  | |||||||||
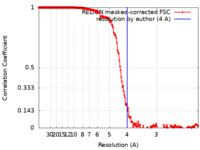
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||
 Authors Authors | Hawkins NC / Kizziah JL | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2021 Journal: Nat Commun / Year: 2021Title: Shape shifter: redirection of prolate phage capsid assembly by staphylococcal pathogenicity islands. Authors: N'Toia C Hawkins / James L Kizziah / José R Penadés / Terje Dokland /   Abstract: Staphylococcus aureus pathogenicity islands (SaPIs) are molecular parasites that hijack helper phages for their transfer. SaPIbov5, the prototypical member of a family of cos type SaPIs, redirects ...Staphylococcus aureus pathogenicity islands (SaPIs) are molecular parasites that hijack helper phages for their transfer. SaPIbov5, the prototypical member of a family of cos type SaPIs, redirects the assembly of ϕ12 helper capsids from prolate to isometric. This size and shape shift is dependent on the SaPIbov5-encoded protein Ccm, a homolog of the ϕ12 capsid protein (CP). Using cryo-electron microscopy, we have determined structures of prolate ϕ12 procapsids and isometric SaPIbov5 procapsids. ϕ12 procapsids have icosahedral end caps with T = 4 architecture and a T = 14 cylindrical midsection, whereas SaPIbov5 procapsids have T = 4 icosahedral architecture. We built atomic models for CP and Ccm, and show that Ccm occupies the pentameric capsomers in the isometric SaPIbov5 procapsids, suggesting that preferential incorporation of Ccm pentamers prevents the cylindrical midsection from forming. Our results highlight that pirate elements have evolved diverse mechanisms to suppress phage multiplication, including the acquisition of phage capsid protein homologs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24720.map.gz emd_24720.map.gz | 470.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24720-v30.xml emd-24720-v30.xml emd-24720.xml emd-24720.xml | 17.5 KB 17.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24720_fsc.xml emd_24720_fsc.xml | 18.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_24720.png emd_24720.png | 181.4 KB | ||
| Masks |  emd_24720_msk_1.map emd_24720_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-24720.cif.gz emd-24720.cif.gz | 6 KB | ||
| Others |  emd_24720_half_map_1.map.gz emd_24720_half_map_1.map.gz emd_24720_half_map_2.map.gz emd_24720_half_map_2.map.gz | 412 MB 412 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24720 http://ftp.pdbj.org/pub/emdb/structures/EMD-24720 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24720 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24720 | HTTPS FTP |
-Validation report
| Summary document |  emd_24720_validation.pdf.gz emd_24720_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_24720_full_validation.pdf.gz emd_24720_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_24720_validation.xml.gz emd_24720_validation.xml.gz | 25.7 KB | Display | |
| Data in CIF |  emd_24720_validation.cif.gz emd_24720_validation.cif.gz | 34.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24720 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24720 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24720 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24720 | HTTPS FTP |
-Related structure data
| Related structure data |  7rwzMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24720.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24720.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.11 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_24720_msk_1.map emd_24720_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_24720_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_24720_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : SaPIbov5 procapsid
| Entire | Name: SaPIbov5 procapsid |
|---|---|
| Components |
|
-Supramolecule #1: SaPIbov5 procapsid
| Supramolecule | Name: SaPIbov5 procapsid / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Cos capsid morphogenesis protein (Ccm)
| Macromolecule | Name: Cos capsid morphogenesis protein (Ccm) / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 40.468102 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKIMKEFKEQ FGYQLSNFDD MDIKGYANLY QKDIGKDVSM IEQGLKQLSI TETEVLLPEQ INKKLLGVLN MNEVNQSSNT WVGTLTKQL VSSENNFNIQ ELPTARKEVF KELLVNRELP QTLRDVITIT DDEHVESIPA LSYIKDKLAT NGIELSLNGS S KYFDRREG ...String: MKIMKEFKEQ FGYQLSNFDD MDIKGYANLY QKDIGKDVSM IEQGLKQLSI TETEVLLPEQ INKKLLGVLN MNEVNQSSNT WVGTLTKQL VSSENNFNIQ ELPTARKEVF KELLVNRELP QTLRDVITIT DDEHVESIPA LSYIKDKLAT NGIELSLNGS S KYFDRREG HIYTEVADSV VHGSDRTLDD LLKEIFINEC VSYETTLLLD KNNASGLIDK DNQDLSLYNQ GIKEVSNTSM YD GIKQAMK DIPQTFRRKV SVVMNTEHHD KLIKELAQMG LGTLAGDLTK LFNVSHVVVT DDAQDIFVGD FGHAIYAKYE PIM YNKKKQ ALKGVYQFAL NYVFDIKIVP ELLRIVKVK UniProtKB: Phage capsid protein |
-Macromolecule #2: Major capsid protein
| Macromolecule | Name: Major capsid protein / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 45.29282 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MRNFKNDNEL LGGNEMPTLY ELKQSLGMIG QQLKNKNDEL SQKATDPNID MEDIKQLETE KAGLQQRFNI VERQVQDIEE KEKAKVKDK GEAYQSLSDN EKMVKAKAEF YRHAILPNEF EKPSMEAQRL LHALPTGNDS GGDKLLPKTL SKEIVSEPFA K NQLREKAR ...String: MRNFKNDNEL LGGNEMPTLY ELKQSLGMIG QQLKNKNDEL SQKATDPNID MEDIKQLETE KAGLQQRFNI VERQVQDIEE KEKAKVKDK GEAYQSLSDN EKMVKAKAEF YRHAILPNEF EKPSMEAQRL LHALPTGNDS GGDKLLPKTL SKEIVSEPFA K NQLREKAR LTNIKGLEIP RVSYTLDDDD FITDVETAKE LKAKGDTVKF TTNKFKVFAA ISDTVIHGSD VDLVNWVENA LQ SGLAAKE RKDALAVSPK SGLEHMSFYN GSVKEVEGAD MYDAIINALA DLHEDYRDNA TIYMRYADYV KIISVLSNGT TNF FDTPAE KVFGKPVVFT DAAVKPIVGD FNYFGINYDG TTYDTDKDVK KGEYLFVLTA WYDQQRTLDS AFRIAKAKEN TGPL PS UniProtKB: ATP-dependent Clp protease proteolytic subunit |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)