[English] 日本語
 Yorodumi
Yorodumi- EMDB-2184: Structure of protozoan virus from the obligate human genitourinar... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-2184 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
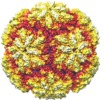
| Title | Structure of protozoan virus from the obligate human genitourinary parasite Trichomonas vaginalis | |||||||||
 Map data Map data | Icosahedral reconstruction of TVV1 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cryo-TEM / image reconstruction / icosahedral reconstruction / Totiviridae / T=2 virus / Trichomonas vaginalis virus 1 | |||||||||
| Biological species |  Trichomonas vaginalis virus Trichomonas vaginalis virus | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 5.5 Å | |||||||||
 Authors Authors | Parent KN / Takagi Y / Cardone G / Olson NH / Ericsson M / Li Y / Fichorova RN / Nibert ML / Baker TS | |||||||||
 Citation Citation |  Journal: mBio / Year: 2013 Journal: mBio / Year: 2013Title: Structure of a protozoan virus from the human genitourinary parasite Trichomonas vaginalis. Authors: Kristin N Parent / Yuko Takagi / Giovanni Cardone / Norman H Olson / Maria Ericsson / May Yang / Yujin Lee / John M Asara / Raina N Fichorova / Timothy S Baker / Max L Nibert /  Abstract: The flagellated protozoan Trichomonas vaginalis is an obligate human genitourinary parasite and the most frequent cause of sexually transmitted disease worldwide. Most clinical isolates of T. ...The flagellated protozoan Trichomonas vaginalis is an obligate human genitourinary parasite and the most frequent cause of sexually transmitted disease worldwide. Most clinical isolates of T. vaginalis are persistently infected with one or more double-stranded RNA (dsRNA) viruses from the genus Trichomonasvirus, family Totiviridae, which appear to influence not only protozoan biology but also human disease. Here we describe the three-dimensional structure of Trichomonas vaginalis virus 1 (TVV1) virions, as determined by electron cryomicroscopy and icosahedral image reconstruction. The structure reveals a T = 1 capsid comprising 120 subunits, 60 in each of two nonequivalent positions, designated A and B, as previously observed for fungal Totiviridae family members. The putative protomer is identified as an asymmetric AB dimer consistent with either decamer or tetramer assembly intermediates. The capsid surface is notable for raised plateaus around the icosahedral 5-fold axes, with canyons connecting the 2- and 3-fold axes. Capsid-spanning channels at the 5-fold axes are unusually wide and may facilitate release of the viral genome, promoting dsRNA-dependent immunoinflammatory responses, as recently shown upon the exposure of human cervicovaginal epithelial cells to either TVV-infected T. vaginalis or purified TVV1 virions. Despite extensive sequence divergence, conservative features of the capsid reveal a helix-rich fold probably derived from an ancestor shared with fungal Totiviridae family members. Also notable are mass spectrometry results assessing the virion proteins as a complement to structure determination, which suggest that translation of the TVV1 RNA-dependent RNA polymerase in fusion with its capsid protein involves -2, and not +1, ribosomal frameshifting, an uncommonly found mechanism to date. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_2184.map.gz emd_2184.map.gz | 109.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-2184-v30.xml emd-2184-v30.xml emd-2184.xml emd-2184.xml | 9.5 KB 9.5 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-2184-tvv_for_EMDB.png EMD-2184-tvv_for_EMDB.png | 139.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-2184 http://ftp.pdbj.org/pub/emdb/structures/EMD-2184 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2184 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-2184 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_2184.map.gz / Format: CCP4 / Size: 468.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_2184.map.gz / Format: CCP4 / Size: 468.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Icosahedral reconstruction of TVV1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.09 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : TVV1 virion
| Entire | Name: TVV1 virion |
|---|---|
| Components |
|
-Supramolecule #1000: TVV1 virion
| Supramolecule | Name: TVV1 virion / type: sample / ID: 1000 / Oligomeric state: icosahedral / Number unique components: 1 |
|---|
-Supramolecule #1: Trichomonas vaginalis virus
| Supramolecule | Name: Trichomonas vaginalis virus / type: virus / ID: 1 / Name.synonym: TVV1 / NCBI-ID: 29256 / Sci species name: Trichomonas vaginalis virus / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No / Syn species name: TVV1 |
|---|---|
| Host (natural) | Organism:  Trichomonas vaginalis (eukaryote) / synonym: PROTOZOA Trichomonas vaginalis (eukaryote) / synonym: PROTOZOA |
| Virus shell | Shell ID: 1 / Diameter: 450 Å / T number (triangulation number): 2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7.6 / Details: 50 mM HEPES, pH 7.2, 0.5M NaCl, 20mM MgCl2 |
| Grid | Details: Quantifoil R2/2 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 99 % / Chamber temperature: 90 K / Instrument: HOMEMADE PLUNGER / Method: blot for 5 sec before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Temperature | Min: 90 K / Max: 90 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at high magnification |
| Date | Apr 26, 2012 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: OTHER / Digitization - Sampling interval: 1.09 µm / Number real images: 84 / Average electron dose: 22 e/Å2 / Bits/pixel: 8 |
| Tilt angle min | 0 |
| Tilt angle max | 0 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 58050 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.3 mm / Nominal defocus max: 4.12 µm / Nominal defocus min: 0.87 µm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: OTHER |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | the particles were selected and preprocessed using RobEM. Image reconstruction was performed using Auto3DEM |
|---|---|
| CTF correction | Details: Robem |
| Final reconstruction | Applied symmetry - Point group: I (icosahedral) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 5.5 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: Auto3DEM / Number images used: 4291 |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















