[English] 日本語
 Yorodumi
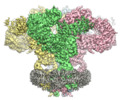
Yorodumi- EMDB-23337: Structure of full-length IP3R1 channel reconstituted into lipid n... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23337 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of full-length IP3R1 channel reconstituted into lipid nanodisc in the apo-state | ||||||||||||||||||||||||
 Map data Map data | Cryo-EM structure of full-length IP3R1 in nanodisc (apo-state) | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
 Keywords Keywords | Calcium channel / lipid nanodisc / MEMBRANE PROTEIN | ||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationEffects of PIP2 hydrolysis / Antigen activates B Cell Receptor (BCR) leading to generation of second messengers / inositol 1,4,5-trisphosphate receptor activity involved in regulation of postsynaptic cytosolic calcium levels / release of sequestered calcium ion into cytosol by endoplasmic reticulum / smooth endoplasmic reticulum membrane / cGMP effects / Elevation of cytosolic Ca2+ levels / platelet dense tubular network / calcineurin complex / platelet dense granule membrane ...Effects of PIP2 hydrolysis / Antigen activates B Cell Receptor (BCR) leading to generation of second messengers / inositol 1,4,5-trisphosphate receptor activity involved in regulation of postsynaptic cytosolic calcium levels / release of sequestered calcium ion into cytosol by endoplasmic reticulum / smooth endoplasmic reticulum membrane / cGMP effects / Elevation of cytosolic Ca2+ levels / platelet dense tubular network / calcineurin complex / platelet dense granule membrane / epithelial fluid transport / inositol 1,4,5-trisphosphate-gated calcium channel activity / phospholipase C-activating G protein-coupled acetylcholine receptor signaling pathway / calcium import into the mitochondrion / voluntary musculoskeletal movement / inositol 1,4,5 trisphosphate binding / positive regulation of hepatocyte proliferation / negative regulation of calcium-mediated signaling / positive regulation of calcium ion transport / endoplasmic reticulum calcium ion homeostasis / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / nuclear inner membrane / Ion homeostasis / transport vesicle membrane / dendrite development / intracellularly gated calcium channel activity / ligand-gated ion channel signaling pathway / intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress / regulation of cytosolic calcium ion concentration / single fertilization / calcium channel inhibitor activity / release of sequestered calcium ion into cytosol / phosphatidylinositol binding / liver regeneration / secretory granule membrane / synaptic membrane / cellular response to cAMP / sarcoplasmic reticulum / post-embryonic development / positive regulation of neuron projection development / positive regulation of insulin secretion / GABA-ergic synapse / Schaffer collateral - CA1 synapse / cell morphogenesis / calcium ion transport / nuclear envelope / presynapse / positive regulation of cytosolic calcium ion concentration / protein phosphatase binding / protein homotetramerization / cellular response to hypoxia / phospholipase C-activating G protein-coupled receptor signaling pathway / transmembrane transporter binding / response to hypoxia / postsynapse / postsynaptic density / positive regulation of apoptotic process / protein domain specific binding / neuronal cell body / calcium ion binding / synapse / dendrite / endoplasmic reticulum membrane / negative regulation of apoptotic process / protein-containing complex binding / nucleolus / perinuclear region of cytoplasm / endoplasmic reticulum / protein homodimerization activity / protein-containing complex / ATP binding / identical protein binding / membrane / plasma membrane / cytoplasm Similarity search - Function | ||||||||||||||||||||||||
| Biological species |  | ||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | ||||||||||||||||||||||||
 Authors Authors | Baker MR / Fan G | ||||||||||||||||||||||||
| Funding support |  United States, 7 items United States, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2021 Journal: Commun Biol / Year: 2021Title: Cryo-EM structure of type 1 IPR channel in a lipid bilayer. Authors: Mariah R Baker / Guizhen Fan / Alexander B Seryshev / Melina A Agosto / Matthew L Baker / Irina I Serysheva /  Abstract: Type 1 inositol 1,4,5-trisphosphate receptor (IPR1) is the predominant Ca-release channel in neurons. IPR1 mediates Ca release from the endoplasmic reticulum into the cytosol and thereby is involved ...Type 1 inositol 1,4,5-trisphosphate receptor (IPR1) is the predominant Ca-release channel in neurons. IPR1 mediates Ca release from the endoplasmic reticulum into the cytosol and thereby is involved in many physiological processes. Here, we present the cryo-EM structures of full-length rat IPR1 reconstituted in lipid nanodisc and detergent solubilized in the presence of phosphatidylcholine determined in ligand-free, closed states by single-particle electron cryo-microscopy. Notably, both structures exhibit the well-established IPR1 protein fold and reveal a nearly complete representation of lipids with similar locations of ordered lipids bound to the transmembrane domains. The lipid-bound structures show improved features that enabled us to unambiguously build atomic models of IPR1 including two membrane associated helices that were not previously resolved in the TM region. Our findings suggest conserved locations of protein-bound lipids among homotetrameric ion channels that are critical for their structural and functional integrity despite the diversity of structural mechanisms for their gating. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23337.map.gz emd_23337.map.gz | 135.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23337-v30.xml emd-23337-v30.xml emd-23337.xml emd-23337.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23337.png emd_23337.png | 266.3 KB | ||
| Filedesc metadata |  emd-23337.cif.gz emd-23337.cif.gz | 7.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23337 http://ftp.pdbj.org/pub/emdb/structures/EMD-23337 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23337 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23337 | HTTPS FTP |
-Validation report
| Summary document |  emd_23337_validation.pdf.gz emd_23337_validation.pdf.gz | 446.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23337_full_validation.pdf.gz emd_23337_full_validation.pdf.gz | 445.7 KB | Display | |
| Data in XML |  emd_23337_validation.xml.gz emd_23337_validation.xml.gz | 7.5 KB | Display | |
| Data in CIF |  emd_23337_validation.cif.gz emd_23337_validation.cif.gz | 8.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23337 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23337 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23337 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23337 | HTTPS FTP |
-Related structure data
| Related structure data |  7lheMC  7lhfC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23337.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23337.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of full-length IP3R1 in nanodisc (apo-state) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Type 1 inositol 1,4,5-trisphosphate receptor tetrameric protein c...
| Entire | Name: Type 1 inositol 1,4,5-trisphosphate receptor tetrameric protein complex |
|---|---|
| Components |
|
-Supramolecule #1: Type 1 inositol 1,4,5-trisphosphate receptor tetrameric protein c...
| Supramolecule | Name: Type 1 inositol 1,4,5-trisphosphate receptor tetrameric protein complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 1.3 MDa |
-Macromolecule #1: Inositol 1,4,5-trisphosphate receptor type 1
| Macromolecule | Name: Inositol 1,4,5-trisphosphate receptor type 1 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 311.84825 KDa |
| Sequence | String: SSFLHIGDIC SLYAEGSTNG FISTLGLVDD RCVVQPEAGD LNNPPKKFRD CLFKLCPMNR YSAQKQFWKA AKPGANSTTD AVLLNKLHH AADLEKKQNE TENRKLLGTV IQYGNVIQLL HLKSNKYLTV NKRLPALLEK NAMRVTLDEA GNEGSWFYIQ P FYKLRSIG ...String: SSFLHIGDIC SLYAEGSTNG FISTLGLVDD RCVVQPEAGD LNNPPKKFRD CLFKLCPMNR YSAQKQFWKA AKPGANSTTD AVLLNKLHH AADLEKKQNE TENRKLLGTV IQYGNVIQLL HLKSNKYLTV NKRLPALLEK NAMRVTLDEA GNEGSWFYIQ P FYKLRSIG DSVVIGDKVV LNPVNAGQPL HASSHQLVDN PGCNEVNSVN CNTSWKIVLF MKWSDNKDDI LKGGDVVRLF HA EQEKFLT CDEHRKKQHV FLRTTGRQSA TSATSSKALW EVEVVQHDPC RGGAGYWNSL FRFKHLATGH YLAAEVDPDF EEE CLEFQP SVDPDQDASR SRLRNAQEKM VYSLVSVPEG NDISSIFELD PTTLRGGDSL VPRNSYVRLR HLCTNTWVHS TNIP IDKEE EKPVMLKIGT SPLKEDKEAF AIVPVSPAEV RDLDFANDAS KVLGSIAGKL EKGTITQNER RSVTKLLEDL VYFVT GGTN SGQDVLEVVF SKPNRERQKL MREQNILKQI FKLLQAPFTD CGDGPMLRLE ELGDQRHAPF RHICRLCYRV LRHSQQ DYR KNQEYIAKQF GFMQKQIGYD VLAEDTITAL LHNNRKLLEK HITAAEIDTF VSLVRKNREP RFLDYLSDLC VSMNKSI PV TQELICKAVL NPTNADILIE TKLVLSRFEF EGVSTGENAL EAGEDEEEVW LFWRDSNKEI RSKSVRELAQ DAKEGQKE D RDVLSYYRYQ LNLFARMCLD RQYLAINEIS GQLDVDLILR CMSDENLPYD LRASFCRLML HMHVDRDPQE QVTPVKYAR LWSEIPSEIA IDDYDSSGAS KDEIKERFAQ TMEFVEEYLR DVVCQRFPFS DKEKNKLTFE VVNLARNLIY FGFYNFSDLL RLTKILLAI LDCVHVTTIF PISKMTKGEE NKGSNVMRSI HGVGELMTQV VLRGGGFLPM TPMAAAPEGN VKQAEPEKED I MVMDTKLK IIEILQFILN VRLDYRISCL LCIFKREFDE SNSQSSETSS GNSSQEGPSN VPGALDFEHI EEQAEGIFGG SE ENTPLDL DDHGGRTFLR VLLHLTMHDY PPLVSGALQL LFRHFSQRQE VLQAFKQVQL LVTSQDVDNY KQIKQDLDQL RSI VEKSEL WVYKGQGPDE PMDGASGENE HKKTEEGTSK PLKHESTSSY NYRVVKEILI RLSKLCVQES ASVRKSRKQQ QRLL RNMGA HAVVLELLQI PYEKAEDTKM QEIMRLAHEF LQNFCAGNQQ NQALLHKHIN LFLNPGILEA VTMQHIFMNN FQLCS EINE RVVQHFVHCI ETHGRNVQYI KFLQTIVKAE GKFIKKCQDM VMAELVNSGE DVLVFYNDRA SFQTLIQMMR SERDRM DEN SPLFMYHIHL VELLAVCTEG KNVYTEIKCN SLLPLDDIVR VVTHEDCIPE VKIAYINFLN HCYVDTEVEM KEIYTSN HM WKLFENFLVD ICRACNNTSD RKHADSVLEK YVTEIVMSIV TTFFSSPFSD QSTTLQTRQP VFVQLLQGVF RVYHCNWL M PSQKASVESC IRVLSDVAKS RAIAIPVDLD SQVNNLFLKS HNIVQKTAMN WRLSARNAAR RDSVLAASRD YRNIIERLQ DIVSALEDRL RPLVQAELSV LVDVLHRPEL LFPENTDARR KCESGGFICK LIKHTKQLLE ENEEKLCIKV LQTLREMMTK DRGYGEKQI SIDELENAEL PQPPEAENST EQELEPSPPL RQLEDHKRGE ALRQILVNRY YGNIRPSGRR ESLTSFGNGP L SPGGPSKP GGGGGGPGSG STSRGEMSLA EVQCHLDKEG ASNLVIDLIM NASSDRVFHE SILLAIALLE GGNTTIQHSF FC RLTEDKK SEKFFKVFYD RMKVAQQEIK ATVTVNTSDL GNKKKDDEVD RDAPSRKKAK EPTTQITEEV RDQLLEASAA TRK AFTTFR READPDDHYQ SGEGTQATTD KAKDDLEMSA VITIMQPILR FLQLLCENHN RDLQNFLRCQ NNKTNYNLVC ETLQ FLDCI CGSTTGGLGL LGLYINEKNV ALINQTLESL TEYCQGPCHE NQNCIATHES NGIDIITALI LNDINPLGKK RMDLV LELK NNASKLLLAI MESRHDSENA ERILYNMRPK ELVEVIKKAY MQGEVEFEDG ENGEDGAASP RNVGHNIYIL AHQLAR HNK ELQTMLKPGG QVDGDEALEF YAKHTAQIEI VRLDRTMEQI VFPVPSICEF LTKESKLRIY YTTERDEQGS KINDFFL RS EDLFNEMNWQ KKLRAQPVLY WCARNMSFWS SISFNLAVLM NLLVAFFYPF KGVRGGTLEP HWSGLLWTAM LISLAIVI A LPKPHGIRAL IASTILRLIF SVGLQPTLFL LGAFNVCNKI IFLMSFVGNC GTFTRGYRAM VLDVEFLYHL LYLLICAMG LFVHEFFYSL LLFDLVYREE TLLNVIKSVT RNGRPIILTA ALALILVYLF SIVGYLFFKD DFILEVDRLP NETAGPETGE SLANDFLYS DVCRVETGEN CTSPAPKEEL LPVEETEQDK EHTCETLLMC IVTVLSHGLR SGGGVGDVLR KPSKEEPLFA A RVIYDLLF FFMVIIIVLN LIFGVIIDTF ADLRSEKQKK EEILKTTCFI CGLERDKFDN KTVTFEEHIK EEHNMWHYLC FI VLVKVKD STEYTGPESY VAEMIRERNL DWFPRMRAMS LVSSDSEGEQ NELRNLQEKL ESTMKLVTNL SGQLSELKDQ MTE QRKQKQ RIGLLGHP UniProtKB: Inositol 1,4,5-trisphosphate-gated calcium channel ITPR1 |
-Macromolecule #2: (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,...
| Macromolecule | Name: (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL type: ligand / ID: 2 / Number of copies: 28 / Formula: PLX |
|---|---|
| Molecular weight | Theoretical: 767.132 Da |
| Chemical component information |  ChemComp-PLX: |
-Macromolecule #3: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 3 / Number of copies: 4 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil / Material: COPPER / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 3840 pixel / Digitization - Dimensions - Height: 3712 pixel / Digitization - Frames/image: 2-35 / Number real images: 22000 / Average exposure time: 0.2 sec. / Average electron dose: 56.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 46943 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)























