[English] 日本語
 Yorodumi
Yorodumi- EMDB-20185: Cryo-EM structure of the N-terminally acetylated C-terminal Alpha... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20185 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the N-terminally acetylated C-terminal Alpha-synuclein truncation Ac1-122 | |||||||||
 Map data Map data | em-volume_P1 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | C-terminal Alpha-synuclein truncation / PROTEIN FIBRIL | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of mitochondrial electron transport, NADH to ubiquinone / : / neutral lipid metabolic process / regulation of acyl-CoA biosynthetic process / negative regulation of dopamine uptake involved in synaptic transmission / negative regulation of norepinephrine uptake / response to desipramine / positive regulation of SNARE complex assembly / positive regulation of hydrogen peroxide catabolic process / supramolecular fiber ...negative regulation of mitochondrial electron transport, NADH to ubiquinone / : / neutral lipid metabolic process / regulation of acyl-CoA biosynthetic process / negative regulation of dopamine uptake involved in synaptic transmission / negative regulation of norepinephrine uptake / response to desipramine / positive regulation of SNARE complex assembly / positive regulation of hydrogen peroxide catabolic process / supramolecular fiber / regulation of synaptic vesicle recycling / negative regulation of chaperone-mediated autophagy / mitochondrial membrane organization / regulation of reactive oxygen species biosynthetic process / positive regulation of protein localization to cell periphery / negative regulation of platelet-derived growth factor receptor signaling pathway / negative regulation of exocytosis / regulation of glutamate secretion / dopamine biosynthetic process / response to iron(II) ion / SNARE complex assembly / negative regulation of dopamine metabolic process / positive regulation of neurotransmitter secretion / regulation of macrophage activation / positive regulation of inositol phosphate biosynthetic process / regulation of norepinephrine uptake / regulation of locomotion / synaptic vesicle transport / negative regulation of microtubule polymerization / transporter regulator activity / synaptic vesicle priming / dopamine uptake involved in synaptic transmission / protein kinase inhibitor activity / regulation of dopamine secretion / negative regulation of thrombin-activated receptor signaling pathway / dynein complex binding / mitochondrial ATP synthesis coupled electron transport / positive regulation of receptor recycling / cuprous ion binding / nuclear outer membrane / response to magnesium ion / positive regulation of exocytosis / synaptic vesicle exocytosis / positive regulation of endocytosis / kinesin binding / synaptic vesicle endocytosis / enzyme inhibitor activity / cysteine-type endopeptidase inhibitor activity / response to type II interferon / negative regulation of serotonin uptake / regulation of presynapse assembly / alpha-tubulin binding / beta-tubulin binding / phospholipase binding / behavioral response to cocaine / supramolecular fiber organization / phospholipid metabolic process / cellular response to fibroblast growth factor stimulus / axon terminus / inclusion body / cellular response to epinephrine stimulus / Hsp70 protein binding / response to interleukin-1 / regulation of microtubule cytoskeleton organization / cellular response to copper ion / positive regulation of release of sequestered calcium ion into cytosol / SNARE binding / adult locomotory behavior / excitatory postsynaptic potential / protein tetramerization / phosphoprotein binding / microglial cell activation / ferrous iron binding / fatty acid metabolic process / regulation of long-term neuronal synaptic plasticity / synapse organization / PKR-mediated signaling / protein destabilization / phospholipid binding / receptor internalization / tau protein binding / long-term synaptic potentiation / terminal bouton / positive regulation of inflammatory response / synaptic vesicle membrane / actin cytoskeleton / growth cone / actin binding / cellular response to oxidative stress / neuron apoptotic process / cell cortex / histone binding / response to lipopolysaccharide / microtubule binding / chemical synaptic transmission / amyloid fibril formation / molecular adaptor activity / negative regulation of neuron apoptotic process / mitochondrial outer membrane / oxidoreductase activity Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
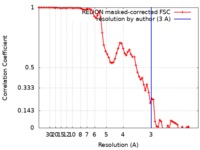
| Method | helical reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Xiaodan N / Ryan PM | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Mol Biol / Year: 2019 Journal: J Mol Biol / Year: 2019Title: Structural Insights into α-Synuclein Fibril Polymorphism: Effects of Parkinson's Disease-Related C-Terminal Truncations. Authors: Xiaodan Ni / Ryan P McGlinchey / Jiansen Jiang / Jennifer C Lee /  Abstract: Lewy bodies, hallmarks of Parkinson's disease, contain C-terminally truncated (ΔC) α-synuclein (α-syn). Here, we report fibril structures of three N-terminally acetylated (Ac) α-syn constructs, ...Lewy bodies, hallmarks of Parkinson's disease, contain C-terminally truncated (ΔC) α-synuclein (α-syn). Here, we report fibril structures of three N-terminally acetylated (Ac) α-syn constructs, Ac1-140, Ac1-122, and Ac1-103, solved by cryoelectron microscopy. Both ΔC-α-syn variants exhibited faster aggregation kinetics, and Ac1-103 fibrils efficiently seeded the full-length protein, highlighting their importance in pathogenesis. Interestingly, fibril helical twists increased upon the removal of C-terminal residues and can be propagated through cross-seeding. Compared to that of Ac1-140, increased electron densities were seen in the N-terminus of Ac1-103, whereas the C-terminus of Ac1-122 appeared more structured. In accord, the respective termini of ΔC-α-syn exhibited increased protease resistance. Despite similar amyloid core residues, distinctive features were seen for both Ac1-122 and Ac1-103. Particularly, Ac1-103 has the tightest packed core with an additional turn, likely attributable to conformational changes in the N-terminal region. These molecular differences offer insights into the effect of C-terminal truncations on α-syn fibril polymorphism. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20185.map.gz emd_20185.map.gz | 27.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20185-v30.xml emd-20185-v30.xml emd-20185.xml emd-20185.xml | 14.1 KB 14.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20185_fsc.xml emd_20185_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_20185.png emd_20185.png | 65.8 KB | ||
| Filedesc metadata |  emd-20185.cif.gz emd-20185.cif.gz | 5.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20185 http://ftp.pdbj.org/pub/emdb/structures/EMD-20185 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20185 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20185 | HTTPS FTP |
-Validation report
| Summary document |  emd_20185_validation.pdf.gz emd_20185_validation.pdf.gz | 593.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_20185_full_validation.pdf.gz emd_20185_full_validation.pdf.gz | 593.2 KB | Display | |
| Data in XML |  emd_20185_validation.xml.gz emd_20185_validation.xml.gz | 9.7 KB | Display | |
| Data in CIF |  emd_20185_validation.cif.gz emd_20185_validation.cif.gz | 12.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20185 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20185 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20185 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-20185 | HTTPS FTP |
-Related structure data
| Related structure data |  6oslMC  6osjC  6osmC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_20185.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20185.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | em-volume_P1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : N-terminally acetylated C-terminal Alpha-synuclein truncation Ac1-122
| Entire | Name: N-terminally acetylated C-terminal Alpha-synuclein truncation Ac1-122 |
|---|---|
| Components |
|
-Supramolecule #1: N-terminally acetylated C-terminal Alpha-synuclein truncation Ac1-122
| Supramolecule | Name: N-terminally acetylated C-terminal Alpha-synuclein truncation Ac1-122 type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Alpha-synuclein
| Macromolecule | Name: Alpha-synuclein / type: protein_or_peptide / ID: 1 / Number of copies: 10 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 14.476108 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDVFMKGLSK AKEGVVAAAE KTKQGVAEAA GKTKEGVLYV GSKTKEGVVH GVATVAEKTK EQVTNVGGAV VTGVTAVAQK TVEGAGSIA AATGFVKKDQ LGKNEEGAPQ EGILEDMPVD PDNEAYEMPS EEGYQDYEPE A UniProtKB: Alpha-synuclein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 1.3 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)