[English] 日本語
 Yorodumi
Yorodumi- EMDB-20729: Cryo-EM structure of the mitochondrial TOM complex from yeast (te... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20729 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the mitochondrial TOM complex from yeast (tetramer) | |||||||||
 Map data Map data | Filtered, sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Membrane protein / mitochondrial protein import / mitochondrial outer membrane / protein translocation / TRANSLOCASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationmitochondrial outer membrane translocase complex assembly / mitochondrial outer membrane translocase complex / protein insertion into mitochondrial outer membrane / protein transmembrane transport / : / porin activity / pore complex / protein import into mitochondrial matrix / protein transmembrane transporter activity / monoatomic ion transport ...mitochondrial outer membrane translocase complex assembly / mitochondrial outer membrane translocase complex / protein insertion into mitochondrial outer membrane / protein transmembrane transport / : / porin activity / pore complex / protein import into mitochondrial matrix / protein transmembrane transporter activity / monoatomic ion transport / mitochondrial intermembrane space / mitochondrial outer membrane / mitochondrion / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
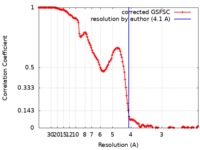
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | |||||||||
 Authors Authors | Park E / Tucker K | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2019 Journal: Nat Struct Mol Biol / Year: 2019Title: Cryo-EM structure of the mitochondrial protein-import channel TOM complex at near-atomic resolution. Authors: Kyle Tucker / Eunyong Park /  Abstract: Nearly all mitochondrial proteins are encoded by the nuclear genome and imported into mitochondria after synthesis on cytosolic ribosomes. These precursor proteins are translocated into mitochondria ...Nearly all mitochondrial proteins are encoded by the nuclear genome and imported into mitochondria after synthesis on cytosolic ribosomes. These precursor proteins are translocated into mitochondria by the TOM complex, a protein-conducting channel in the mitochondrial outer membrane. We have determined high-resolution cryo-EM structures of the core TOM complex from Saccharomyces cerevisiae in dimeric and tetrameric forms. Dimeric TOM consists of two copies each of five proteins arranged in two-fold symmetry: pore-forming β-barrel protein Tom40 and four auxiliary α-helical transmembrane proteins. The pore of each Tom40 has an overall negatively charged inner surface attributed to multiple functionally important acidic patches. The tetrameric complex is essentially a dimer of dimeric TOM, which may be capable of forming higher-order oligomers. Our study reveals the detailed molecular organization of the TOM complex and provides new insights about the mechanism of protein translocation into mitochondria. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20729.map.gz emd_20729.map.gz | 229.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20729-v30.xml emd-20729-v30.xml emd-20729.xml emd-20729.xml | 18.8 KB 18.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_20729_fsc.xml emd_20729_fsc.xml | 14.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_20729.png emd_20729.png | 181.6 KB | ||
| Filedesc metadata |  emd-20729.cif.gz emd-20729.cif.gz | 6 KB | ||
| Others |  emd_20729_additional.map.gz emd_20729_additional.map.gz | 226.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20729 http://ftp.pdbj.org/pub/emdb/structures/EMD-20729 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20729 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20729 | HTTPS FTP |
-Related structure data
| Related structure data |  6ucvMC  6ucuC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20729.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20729.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
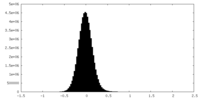
| Annotation | Filtered, sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.15 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Raw combined map
| File | emd_20729_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
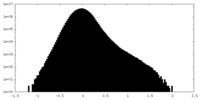
| Annotation | Raw combined map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Tetrameric TOM complex from yeast
| Entire | Name: Tetrameric TOM complex from yeast |
|---|---|
| Components |
|
-Supramolecule #1: Tetrameric TOM complex from yeast
| Supramolecule | Name: Tetrameric TOM complex from yeast / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
-Macromolecule #1: Mitochondrial import receptor subunit TOM40
| Macromolecule | Name: Mitochondrial import receptor subunit TOM40 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 43.227387 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSAPTPLAEA SQIPTIPALS PLTAKQSKGN FFSSNPISSF VVDTYKQLHS HRQSLELVNP GTVENLNKEV SRDVFLSQYF FTGLRADLN KAFSMNPAFQ TSHTFSIGSQ ALPKYAFSAL FANDNLFAQG NIDNDLSVSG RLNYGWDKKN ISKVNLQISD G QPTMCQLE ...String: MSAPTPLAEA SQIPTIPALS PLTAKQSKGN FFSSNPISSF VVDTYKQLHS HRQSLELVNP GTVENLNKEV SRDVFLSQYF FTGLRADLN KAFSMNPAFQ TSHTFSIGSQ ALPKYAFSAL FANDNLFAQG NIDNDLSVSG RLNYGWDKKN ISKVNLQISD G QPTMCQLE QDYQASDFSV NVKTLNPSFS EKGEFTGVAV ASFLQSVTPQ LALGLETLYS RTDGSAPGDA GVSYLTRYVS KK QDWIFSG QLQANGALIA SLWRKVAQNV EAGIETTLQA GMVPITDPLM GTPIGIQPTV EGSTTIGAKY EYRQSVYRGT LDS NGKVAC FLERKVLPTL SVLFCGEIDH FKNDTKIGCG LQFETAGNQE LLMLQQGLDA DGNPLQALPQ LGGWSHPQFE K UniProtKB: Mitochondrial import receptor subunit TOM40 |
-Macromolecule #2: Mitochondrial import receptor subunit TOM22
| Macromolecule | Name: Mitochondrial import receptor subunit TOM22 / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 18.02065 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MVELTEIKDD VVQLDEPQFS RNQAIVEEKA SATNNDVVDD EDDSDSDFED EFDENETLLD RIVALKDIVP PGKRQTISNF FGFTSSFVR NAFTKSGNLA WTLTTTALLL GVPLSLSILA EQQLIEMEKT FDLQSDANNI LAQGEKDAAA TANGGHHHHH H HH UniProtKB: Mitochondrial import receptor subunit TOM22 |
-Macromolecule #3: Mitochondrial import receptor subunit TOM5
| Macromolecule | Name: Mitochondrial import receptor subunit TOM5 / type: protein_or_peptide / ID: 3 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 5.993924 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MFGLPQQEVS EEEKRAHQEQ TEKTLKQAAY VAAFLWVSPM IWHLVKKQWK UniProtKB: Mitochondrial import receptor subunit TOM5 |
-Macromolecule #4: Mitochondrial import receptor subunit TOM6
| Macromolecule | Name: Mitochondrial import receptor subunit TOM6 / type: protein_or_peptide / ID: 4 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 6.41046 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDGMFAMPGA AAGAASPQQP KSRFQAFKES PLYTIALNGA FFVAGVAFIQ SPLMDMLAPQ L UniProtKB: Mitochondrial import receptor subunit TOM6 |
-Macromolecule #5: Mitochondrial import receptor subunit TOM7
| Macromolecule | Name: Mitochondrial import receptor subunit TOM7 / type: protein_or_peptide / ID: 5 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 6.876955 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSFLPSFILS DESKERISKI LTLTHNVAHY GWIPFVLYLG WAHTSNRPNF LNLLSPLPSV UniProtKB: Mitochondrial import receptor subunit TOM7 |
-Macromolecule #6: 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE
| Macromolecule | Name: 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE / type: ligand / ID: 6 / Number of copies: 2 / Formula: MC3 |
|---|---|
| Molecular weight | Theoretical: 677.933 Da |
| Chemical component information | 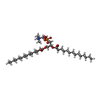 ChemComp-MC3: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Support film - Material: CARBON / Support film - topology: HOLEY / Details: unspecified |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: COUNTING / Average electron dose: 43.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)