+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12875 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The archaellum of Methanocaldococcus villosus | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Archaellum / archaellin / Methanocaldococcus villosus / cryoEM / helical reconstruction / PROTEIN FIBRIL | |||||||||
| Biological species |  Methanocaldococcus villosus (archaea) Methanocaldococcus villosus (archaea) | |||||||||
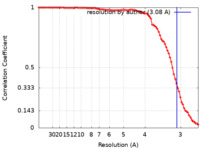
| Method | helical reconstruction / cryo EM / Resolution: 3.08 Å | |||||||||
 Authors Authors | Isupov M / Gambelli L | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2022 Journal: Nat Commun / Year: 2022Title: An archaellum filament composed of two alternating subunits. Authors: Lavinia Gambelli / Michail N Isupov / Rebecca Conners / Mathew McLaren / Annett Bellack / Vicki Gold / Reinhard Rachel / Bertram Daum /   Abstract: Archaea use a molecular machine, called the archaellum, to swim. The archaellum consists of an ATP-powered intracellular motor that drives the rotation of an extracellular filament composed of ...Archaea use a molecular machine, called the archaellum, to swim. The archaellum consists of an ATP-powered intracellular motor that drives the rotation of an extracellular filament composed of multiple copies of proteins named archaellins. In many species, several archaellin homologs are encoded in the same operon; however, previous structural studies indicated that archaellum filaments mainly consist of only one protein species. Here, we use electron cryo-microscopy to elucidate the structure of the archaellum from Methanocaldococcus villosus at 3.08 Å resolution. The filament is composed of two alternating archaellins, suggesting that the architecture and assembly of archaella is more complex than previously thought. Moreover, we identify structural elements that may contribute to the filament's flexibility. #1:  Journal: Faraday Disc.Chem.Soc / Year: 2022 Journal: Faraday Disc.Chem.Soc / Year: 2022Title: Escaping the symmetry trap in helical reconstruction Authors: Gambelli L / Isupov MN / Daum B | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12875.map.gz emd_12875.map.gz | 57.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12875-v30.xml emd-12875-v30.xml emd-12875.xml emd-12875.xml | 19.1 KB 19.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12875_fsc.xml emd_12875_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_12875.png emd_12875.png | 163.6 KB | ||
| Filedesc metadata |  emd-12875.cif.gz emd-12875.cif.gz | 6.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12875 http://ftp.pdbj.org/pub/emdb/structures/EMD-12875 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12875 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12875 | HTTPS FTP |
-Related structure data
| Related structure data |  7ofqMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10884 (Title: New insights into the architecture and dynamics of archaella EMPIAR-10884 (Title: New insights into the architecture and dynamics of archaellaData size: 3.3 TB Data #1: Unaligned 39-frames movies of Methanocaldococcus villosus archaella [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_12875.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12875.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.39 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Archaellum
| Entire | Name: Archaellum |
|---|---|
| Components |
|
-Supramolecule #1: Archaellum
| Supramolecule | Name: Archaellum / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Methanocaldococcus villosus (archaea) Methanocaldococcus villosus (archaea) |
-Macromolecule #1: Archaellin
| Macromolecule | Name: Archaellin / type: protein_or_peptide / ID: 1 / Number of copies: 23 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Methanocaldococcus villosus (archaea) Methanocaldococcus villosus (archaea) |
| Molecular weight | Theoretical: 22.084279 KDa |
| Recombinant expression | Organism:  Methanocaldococcus villosus (archaea) Methanocaldococcus villosus (archaea) |
| Sequence | String: AIGIGTLIIF IAMVLVAAVA AAVLINTSGF LQQKAMATGK ESTEQVASGL LCSGVTGHYV KNKGIDRIVI YITPNAGSAP IDLKQCKLF LMYDGKAVSL NFSKYDTNTV GDFTNGIKDI FNTTVVKWNN ADATSFVVVA LQDDDKSLLT NAVINKGDLA G VLVNVSAA ...String: AIGIGTLIIF IAMVLVAAVA AAVLINTSGF LQQKAMATGK ESTEQVASGL LCSGVTGHYV KNKGIDRIVI YITPNAGSAP IDLKQCKLF LMYDGKAVSL NFSKYDTNTV GDFTNGIKDI FNTTVVKWNN ADATSFVVVA LQDDDKSLLT NAVINKGDLA G VLVNVSAA FGKHVGTRER VSGYLQPEFG APAVIEFTTP AAFTSDVIEL Q |
-Macromolecule #2: Archaellin
| Macromolecule | Name: Archaellin / type: protein_or_peptide / ID: 2 / Number of copies: 22 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Methanocaldococcus villosus (archaea) Methanocaldococcus villosus (archaea) |
| Molecular weight | Theoretical: 22.273643 KDa |
| Recombinant expression | Organism:  Methanocaldococcus villosus (archaea) Methanocaldococcus villosus (archaea) |
| Sequence | String: AIGIGTLIIF IAMVLVAAVA AAVLINTSGF LQQKAMATGK ESTEQVASGL QVIRVLGNHS GGKINWLAVL ISPNAGSAPI DLSQATVMI TDGTHKVIAK YNSTFFNGTL KNGGSIFEAK YNNTTALKPL FDDLPATAFG IVVLQDADTS CSKDTPVINK G DIVAICLN ...String: AIGIGTLIIF IAMVLVAAVA AAVLINTSGF LQQKAMATGK ESTEQVASGL QVIRVLGNHS GGKINWLAVL ISPNAGSAPI DLSQATVMI TDGTHKVIAK YNSTFFNGTL KNGGSIFEAK YNNTTALKPL FDDLPATAFG IVVLQDADTS CSKDTPVINK G DIVAICLN VSNTLNLKPR TKVTGAVIPE FGAPAVISFT TPATYLDTQH IIELQ |
-Macromolecule #3: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 3 / Number of copies: 45 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #4: UNKNOWN LIGAND
| Macromolecule | Name: UNKNOWN LIGAND / type: ligand / ID: 4 / Number of copies: 804 / Formula: UNL |
|---|---|
| Chemical component information | 
ChemComp-UNL: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Buffer | pH: 7 / Component - Concentration: 5.0 mM / Component - Formula: C8H18N2O4S / Component - Name: HEPES |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 40 sec. / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 294 K / Instrument: FEI VITROBOT MARK III / Details: blotting time of 4 sec, -1 blot force. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Number grids imaged: 1 / Number real images: 2795 / Average exposure time: 1.0 sec. / Average electron dose: 37.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: SPOT SCAN / Imaging mode: OTHER / Cs: 2.7 mm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)