+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0297 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Potato virus Y | ||||||||||||
 Map data Map data | Final cryo-EM map of Potato virus Y. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | plant virus / potyvirus / flexible filamentous particle / helical symmetry / VIRUS | ||||||||||||
| Function / homology | Potyvirus coat protein / Potyvirus coat protein / viral capsid / Genome polyprotein Function and homology information Function and homology information | ||||||||||||
| Biological species |  Potato virus Y Potato virus Y | ||||||||||||
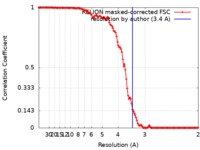
| Method | helical reconstruction / cryo EM / Resolution: 3.4 Å | ||||||||||||
 Authors Authors | Podobnik M / Kezar A | ||||||||||||
| Funding support |  Slovenia, Slovenia,  Czech Republic, 3 items Czech Republic, 3 items
| ||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2019 Journal: Sci Adv / Year: 2019Title: Structural basis for the multitasking nature of the potato virus Y coat protein. Authors: Andreja Kežar / Luka Kavčič / Martin Polák / Jiří Nováček / Ion Gutiérrez-Aguirre / Magda Tušek Žnidarič / Anna Coll / Katja Stare / Kristina Gruden / Maja Ravnikar / David ...Authors: Andreja Kežar / Luka Kavčič / Martin Polák / Jiří Nováček / Ion Gutiérrez-Aguirre / Magda Tušek Žnidarič / Anna Coll / Katja Stare / Kristina Gruden / Maja Ravnikar / David Pahovnik / Ema Žagar / Franci Merzel / Gregor Anderluh / Marjetka Podobnik /   Abstract: Potato virus Y (PVY) is among the most economically important plant pathogens. Using cryoelectron microscopy, we determined the near-atomic structure of PVY's flexuous virions, revealing a previously ...Potato virus Y (PVY) is among the most economically important plant pathogens. Using cryoelectron microscopy, we determined the near-atomic structure of PVY's flexuous virions, revealing a previously unknown lumenal interplay between extended carboxyl-terminal regions of the coat protein units and viral RNA. RNA-coat protein interactions are crucial for the helical configuration and stability of the virion, as revealed by the unique near-atomic structure of RNA-free virus-like particles. The structures offer the first evidence for plasticity of the coat protein's amino- and carboxyl-terminal regions. Together with mutational analysis and in planta experiments, we show their crucial role in PVY infectivity and explain the ability of the coat protein to perform multiple biological tasks. Moreover, the high modularity of PVY virus-like particles suggests their potential as a new molecular scaffold for nanobiotechnological applications. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0297.map.gz emd_0297.map.gz | 17 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0297-v30.xml emd-0297-v30.xml emd-0297.xml emd-0297.xml | 19.2 KB 19.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0297_fsc.xml emd_0297_fsc.xml | 12.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_0297.png emd_0297.png | 101.7 KB | ||
| Filedesc metadata |  emd-0297.cif.gz emd-0297.cif.gz | 6.4 KB | ||
| Others |  emd_0297_half_map_1.map.gz emd_0297_half_map_1.map.gz emd_0297_half_map_2.map.gz emd_0297_half_map_2.map.gz | 140.3 MB 140.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0297 http://ftp.pdbj.org/pub/emdb/structures/EMD-0297 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0297 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0297 | HTTPS FTP |
-Related structure data
| Related structure data |  6hxxMC  0298C  6hxzC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0297.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0297.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final cryo-EM map of Potato virus Y. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.061 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: First half-map after 3D auto-refine in Relion.
| File | emd_0297_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | First half-map after 3D auto-refine in Relion. | ||||||||||||
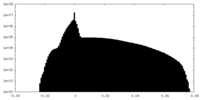
| Projections & Slices |
| ||||||||||||
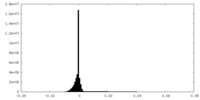
| Density Histograms |
-Half map: Second half-map after 3D auto-refine in Relion.
| File | emd_0297_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Second half-map after 3D auto-refine in Relion. | ||||||||||||
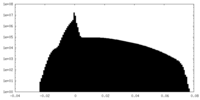
| Projections & Slices |
| ||||||||||||
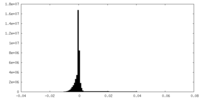
| Density Histograms |
- Sample components
Sample components
-Entire : Potato virus Y
| Entire | Name:  Potato virus Y Potato virus Y |
|---|---|
| Components |
|
-Supramolecule #1: Potato virus Y
| Supramolecule | Name: Potato virus Y / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / Details: Potato virus Y / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  |
-Supramolecule #2: Coat protein
| Supramolecule | Name: Coat protein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 Details: Approximately 2000 copies of PVY coat protein assemble around viral ssRNA to form the virion. |
|---|---|
| Source (natural) | Organism:  Potato virus Y / Strain: NTN Potato virus Y / Strain: NTN |
| Molecular weight | Theoretical: 299 KDa |
-Supramolecule #3: Viral RNA
| Supramolecule | Name: Viral RNA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 Details: Viral genome consists of positive-sense single stranded RNA. |
|---|---|
| Source (natural) | Organism:  Potato virus Y / Strain: NTN Potato virus Y / Strain: NTN |
-Macromolecule #1: Coat protein
| Macromolecule | Name: Coat protein / type: protein_or_peptide / ID: 1 / Number of copies: 35 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Potato virus Y Potato virus Y |
| Molecular weight | Theoretical: 29.935797 KDa |
| Sequence | String: GNDTIDAGGS TKKDAKQEQG SIQPNLNKEK EKDVNVGTSG THTVPRIKAI TSKMRMPKSK GATVLNLEHL LEYAPQQIDI SNTRATQSQ FDTWYEAVQL AYDIGETEMP TVMNGLMVWC IENGTSPNIN GVWVMMDGDE QVEYPLKPIV ENAKPTLRQI M AHFSDVAE ...String: GNDTIDAGGS TKKDAKQEQG SIQPNLNKEK EKDVNVGTSG THTVPRIKAI TSKMRMPKSK GATVLNLEHL LEYAPQQIDI SNTRATQSQ FDTWYEAVQL AYDIGETEMP TVMNGLMVWC IENGTSPNIN GVWVMMDGDE QVEYPLKPIV ENAKPTLRQI M AHFSDVAE AYIEMRNKKE PYMPRYGLVR NLRDGSLARY AFDFYEVTSR TPVRAREAHI QMKAAALKSA QSRLFGLDGG IS TQEENTE RHTTEDVSPS MHTLLGVKNM UniProtKB: Genome polyprotein |
-Macromolecule #2: RNA (5'-R(P*UP*UP*UP*UP*U)-3')
| Macromolecule | Name: RNA (5'-R(P*UP*UP*UP*UP*U)-3') / type: rna / ID: 2 Details: Due to helical averaging during cryo-EM data processing, all nucleotides were assigned the uracil base. Number of copies: 35 |
|---|---|
| Source (natural) | Organism:  Potato virus Y Potato virus Y |
| Molecular weight | Theoretical: 1.485872 KDa |
| Sequence | String: UUUUU |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | helical array |
- Sample preparation
Sample preparation
| Concentration | 0.9 mg/mL |
|---|---|
| Buffer | pH: 8 / Component - Concentration: 5.0 mM / Component - Formula: B4Na2O7 / Component - Name: disodium tetraborate |
| Grid | Model: Quantifoil R2/1 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Frames/image: 1-16 / Number real images: 726 / Average electron dose: 48.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 75000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)