[English] 日本語
 Yorodumi
Yorodumi- EMDB-11418: E. coli 70S-RNAP expressome complex in NusG-coupled state (38 nt ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-11418 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | E. coli 70S-RNAP expressome complex in NusG-coupled state (38 nt intervening mRNA) | ||||||||||||||||||
 Map data Map data | NusG-coupled E. coli expressome with intervening mRNA length 38 nucleotides: composite map. | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | Transcription / Translation / Expressome / Ribosome / RNA polymerase / GENE REGULATION | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of cytoplasmic translation / transcription elongation-coupled chromatin remodeling / RNA polymerase complex / stringent response / submerged biofilm formation / cellular response to cell envelope stress / regulation of DNA-templated transcription initiation / bacterial-type flagellum assembly / bacterial-type RNA polymerase core enzyme binding / cytosolic DNA-directed RNA polymerase complex ...positive regulation of cytoplasmic translation / transcription elongation-coupled chromatin remodeling / RNA polymerase complex / stringent response / submerged biofilm formation / cellular response to cell envelope stress / regulation of DNA-templated transcription initiation / bacterial-type flagellum assembly / bacterial-type RNA polymerase core enzyme binding / cytosolic DNA-directed RNA polymerase complex / bacterial-type flagellum-dependent cell motility / transcriptional attenuation / endoribonuclease inhibitor activity / positive regulation of ribosome biogenesis / RNA-binding transcription regulator activity / nitrate assimilation / translational termination / negative regulation of cytoplasmic translation / DnaA-L2 complex / translation repressor activity / negative regulation of DNA-templated DNA replication initiation / DNA-directed RNA polymerase complex / mRNA regulatory element binding translation repressor activity / assembly of large subunit precursor of preribosome / cytosolic ribosome assembly / response to reactive oxygen species / regulation of DNA-templated transcription elongation / ribosome assembly / transcription elongation factor complex / transcription antitermination / regulation of cell growth / cell motility / DNA-templated transcription initiation / DNA-templated transcription termination / response to radiation / mRNA 5'-UTR binding / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / large ribosomal subunit / ribosome biogenesis / transferase activity / ribosome binding / response to heat / ribosomal small subunit biogenesis / ribosomal small subunit assembly / protein-containing complex assembly / 5S rRNA binding / ribosomal large subunit assembly / small ribosomal subunit / small ribosomal subunit rRNA binding / large ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / cytosolic large ribosomal subunit / intracellular iron ion homeostasis / cytoplasmic translation / tRNA binding / protein dimerization activity / single-stranded RNA binding / negative regulation of translation / rRNA binding / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / response to antibiotic / negative regulation of DNA-templated transcription / mRNA binding / DNA-templated transcription / magnesium ion binding / DNA binding / RNA binding / zinc ion binding / metal ion binding / membrane / cytosol / cytoplasm Similarity search - Function | ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
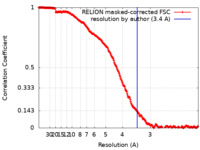
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | ||||||||||||||||||
 Authors Authors | Webster MW / Takacs M | ||||||||||||||||||
| Funding support |  France, 5 items France, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Science / Year: 2020 Journal: Science / Year: 2020Title: Structural basis of transcription-translation coupling and collision in bacteria. Authors: Michael William Webster / Maria Takacs / Chengjin Zhu / Vita Vidmar / Ayesha Eduljee / Mo'men Abdelkareem / Albert Weixlbaumer /  Abstract: Prokaryotic messenger RNAs (mRNAs) are translated as they are transcribed. The lead ribosome potentially contacts RNA polymerase (RNAP) and forms a supramolecular complex known as the expressome. The ...Prokaryotic messenger RNAs (mRNAs) are translated as they are transcribed. The lead ribosome potentially contacts RNA polymerase (RNAP) and forms a supramolecular complex known as the expressome. The basis of expressome assembly and its consequences for transcription and translation are poorly understood. Here, we present a series of structures representing uncoupled, coupled, and collided expressome states determined by cryo-electron microscopy. A bridge between the ribosome and RNAP can be formed by the transcription factor NusG, which stabilizes an otherwise-variable interaction interface. Shortening of the intervening mRNA causes a substantial rearrangement that aligns the ribosome entrance channel to the RNAP exit channel. In this collided complex, NusG linkage is no longer possible. These structures reveal mechanisms of coordination between transcription and translation and provide a framework for future study. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
-Validation report
| Summary document |  emd_11418_validation.pdf.gz emd_11418_validation.pdf.gz | 164.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_11418_full_validation.pdf.gz emd_11418_full_validation.pdf.gz | 164.1 KB | Display | |
| Data in XML |  emd_11418_validation.xml.gz emd_11418_validation.xml.gz | 572 B | Display | |
| Data in CIF |  emd_11418_validation.cif.gz emd_11418_validation.cif.gz | 482 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11418 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11418 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11418 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-11418 | HTTPS FTP |
-Related structure data
| Related structure data |  6ztjMC  6ztlC  6ztmC  6ztnC  6ztoC  6ztpC  6zu1C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_11418.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_11418.map.gz / Format: CCP4 / Size: 1000 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | NusG-coupled E. coli expressome with intervening mRNA length 38 nucleotides: composite map. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
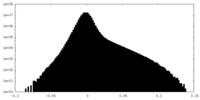
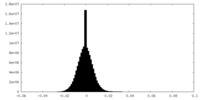
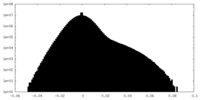
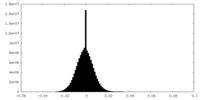
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.052 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
+Additional map: NusG-coupled E. coli expressome with intervening mRNA length...
+Additional map: NusG-coupled E. coli expressome with intervening mRNA length...
+Additional map: NusG-coupled E. coli expressome with intervening mRNA length...
+Additional map: NusG-coupled E. coli expressome with intervening mRNA length...
+Additional map: NusG-coupled E. coli expressome with intervening mRNA length...
+Additional map: NusG-coupled E. coli expressome with intervening mRNA length...
+Additional map: NusG-coupled E. coli expressome with intervening mRNA length...
+Additional map: NusG-coupled E. coli expressome with intervening mRNA length...
+Additional map: NusG-coupled E. coli expressome with intervening mRNA length...
+Additional map: NusG-coupled E. coli expressome with intervening mRNA length...
+Additional map: NusG-coupled E. coli expressome with intervening mRNA length...
+Additional map: NusG-coupled E. coli expressome with intervening mRNA length...
+Additional map: NusG-coupled E. coli expressome with intervening mRNA length...
+Additional map: NusG-coupled E. coli expressome with intervening mRNA length...
+Additional map: NusG-coupled E. coli expressome with intervening mRNA length...
+Additional map: NusG-coupled E. coli expressome with intervening mRNA length...
+Half map: NusG-coupled E. coli expressome with intervening mRNA length...
+Half map: NusG-coupled E. coli expressome with intervening mRNA length...
- Sample components
Sample components
+Entire : E. coli 70S-RNAP expressome complex in NusG-coupled state
+Supramolecule #1: E. coli 70S-RNAP expressome complex in NusG-coupled state
+Supramolecule #2: 70S ribosome within the NusG-coupled expressome
+Supramolecule #3: DNA-directed RNA polymerase within the NusG-coupled expressome
+Supramolecule #4: synthetic mRNA and template DNA
+Macromolecule #1: 16S ribosomal RNA
+Macromolecule #22: mRNA
+Macromolecule #23: tRNA(fmet) P-site
+Macromolecule #24: Phe-NH-tRNA(Phe) A-site
+Macromolecule #26: 23S ribosomal RNA
+Macromolecule #27: 5S ribosomal RNA
+Macromolecule #2: 30S ribosomal protein S2
+Macromolecule #3: 30S ribosomal protein S3
+Macromolecule #4: 30S ribosomal protein S4
+Macromolecule #5: 30S ribosomal protein S5
+Macromolecule #6: 30S ribosomal protein S6
+Macromolecule #7: 30S ribosomal protein S7
+Macromolecule #8: 30S ribosomal protein S8
+Macromolecule #9: 30S ribosomal protein S9
+Macromolecule #10: 30S ribosomal protein S10
+Macromolecule #11: 30S ribosomal protein S11
+Macromolecule #12: 30S ribosomal protein S12
+Macromolecule #13: 30S ribosomal protein S13
+Macromolecule #14: 30S ribosomal protein S14
+Macromolecule #15: 30S ribosomal protein S15
+Macromolecule #16: 30S ribosomal protein S16
+Macromolecule #17: 30S ribosomal protein S17
+Macromolecule #18: 30S ribosomal protein S18
+Macromolecule #19: 30S ribosomal protein S19
+Macromolecule #20: 30S ribosomal protein S20
+Macromolecule #21: 30S ribosomal protein S21
+Macromolecule #25: 30S ribosomal protein S1
+Macromolecule #28: 50S ribosomal protein L2
+Macromolecule #29: 50S ribosomal protein L3
+Macromolecule #30: 50S ribosomal protein L4
+Macromolecule #31: 50S ribosomal protein L5
+Macromolecule #32: 50S ribosomal protein L6
+Macromolecule #33: 50S ribosomal protein L9
+Macromolecule #34: 50S ribosomal protein L10
+Macromolecule #35: 50S ribosomal protein L11
+Macromolecule #36: 50S ribosomal protein L13
+Macromolecule #37: 50S ribosomal protein L14
+Macromolecule #38: 50S ribosomal protein L15
+Macromolecule #39: 50S ribosomal protein L16
+Macromolecule #40: 50S ribosomal protein L17
+Macromolecule #41: 50S ribosomal protein L18
+Macromolecule #42: 50S ribosomal protein L19
+Macromolecule #43: 50S ribosomal protein L20
+Macromolecule #44: 50S ribosomal protein L21
+Macromolecule #45: 50S ribosomal protein L22
+Macromolecule #46: 50S ribosomal protein L23
+Macromolecule #47: 50S ribosomal protein L24
+Macromolecule #48: 50S ribosomal protein L25
+Macromolecule #49: 50S ribosomal protein L27
+Macromolecule #50: 50S ribosomal protein L28
+Macromolecule #51: 50S ribosomal protein L29
+Macromolecule #52: 50S ribosomal protein L30
+Macromolecule #53: 50S ribosomal protein L32
+Macromolecule #54: 50S ribosomal protein L33
+Macromolecule #55: 50S ribosomal protein L34
+Macromolecule #56: 50S ribosomal protein L35
+Macromolecule #57: 50S ribosomal protein L36
+Macromolecule #58: 50S ribosomal protein L31
+Macromolecule #61: DNA-directed RNA polymerase subunit alpha
+Macromolecule #62: DNA-directed RNA polymerase subunit beta
+Macromolecule #63: DNA-directed RNA polymerase subunit beta'
+Macromolecule #64: DNA-directed RNA polymerase subunit omega
+Macromolecule #65: Transcription termination/antitermination protein NusG
+Macromolecule #59: Non-template DNA strand
+Macromolecule #60: Template DNA strand
+Macromolecule #66: MAGNESIUM ION
+Macromolecule #67: PHENYLALANINE
+Macromolecule #68: ZINC ION
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
| |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 42.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.5 µm / Nominal defocus min: 0.7000000000000001 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Correlation coefficient | ||||||
| Output model |  PDB-6ztj: |
 Movie
Movie Controller
Controller
























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)