+検索条件
-Structure paper
| タイトル | Crystallographic Fragment Screening of an Entire Library |
|---|---|
| ジャーナル・号・ページ | To Be Published |
| 掲載日 | 2015年2月10日 (構造データの登録日) |
 著者 著者 | Heine, A. / Knoerlein, A. / Schiebel, J. / Klebe, G. |
 リンク リンク | PubMedで検索 |
| 手法 | X線回折 |
| 解像度 | 0.99 - 1.754 Å |
| 構造データ |  PDB-4y35:  PDB-4y36:  PDB-4y37:  PDB-4y39:  PDB-4y3e:  PDB-4y3f:  PDB-4y3g:  PDB-4y3m:  PDB-4y3n:  PDB-4y3p:  PDB-4y3q:  PDB-4y3r:  PDB-4y3s:  PDB-4y3w:  PDB-4y3x:  PDB-4y3z:  PDB-4y41:  PDB-4y43:  PDB-4y44:  PDB-4y45:  PDB-4y47:  PDB-4y4a:  PDB-4y4b:  PDB-4y4e:  PDB-4y4t:  PDB-4y4u:  PDB-4y4w:  PDB-4y4x:  PDB-4y4z:  PDB-4y50:  PDB-4y51:  PDB-4y53:  PDB-4y54:  PDB-4y56:  PDB-4y57:  PDB-4y5l:  PDB-4y5m:  PDB-4y5n:  PDB-4y5p:  PDB-4yck:  PDB-4yct:  PDB-4ycy:  PDB-4yd3:  PDB-4yd4:  PDB-4yd5:  PDB-4yd6:  PDB-4yd7:  PDB-5dpz:  PDB-5dq1:  PDB-5dq2:  PDB-5dq4:  PDB-5dq5:  PDB-5dr0:  PDB-5dr1:  PDB-5dr3:  PDB-5dr4:  PDB-5dr7:  PDB-5dr8:  PDB-5hco:  PDB-5is4:  PDB-5isj:  PDB-5isk:  PDB-5j25: |
| 化合物 |  ChemComp-ACT:  ChemComp-GOL:  ChemComp-DMS:  ChemComp-F90:  ChemComp-HOH:  ChemComp-EDO:  ChemComp-PGE:  ChemComp-45L:  ChemComp-45J:  ChemComp-PEG:  ChemComp-45M:  ChemComp-F05: 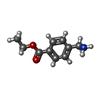 ChemComp-78B: 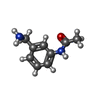 ChemComp-463: 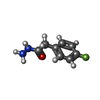 ChemComp-45Y: 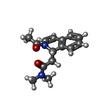 ChemComp-45V:  ChemComp-46S: 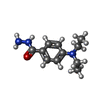 ChemComp-F02: 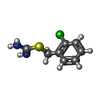 ChemComp-F06: 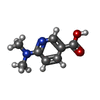 ChemComp-46L:  ChemComp-46Q: 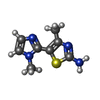 ChemComp-46P: 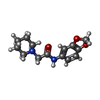 ChemComp-F41: 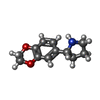 ChemComp-46O: 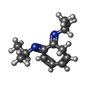 ChemComp-46T: 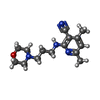 ChemComp-46Y:  ChemComp-F91: 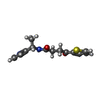 ChemComp-479: 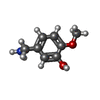 ChemComp-47G: 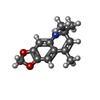 ChemComp-47H: 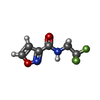 ChemComp-46R:  ChemComp-46X:  ChemComp-46V:  ChemComp-SO4:  ChemComp-46W:  ChemComp-47Q:  ChemComp-47R: 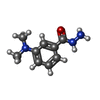 ChemComp-47S:  ChemComp-47A: 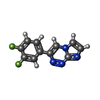 ChemComp-47B:  ChemComp-47E: 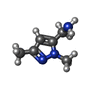 ChemComp-47J: 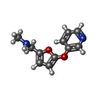 ChemComp-F63:  ChemComp-PG4:  ChemComp-NA:  ChemComp-47Y:  ChemComp-487:  ChemComp-483:  ChemComp-FBF:  ChemComp-4AO:  ChemComp-4AQ:  ChemComp-1PE:  ChemComp-4AY:  ChemComp-BR:  ChemComp-4AV:  ChemComp-4AS:  ChemComp-F5N:  ChemComp-51V:  ChemComp-5HL:  ChemComp-5HO:  ChemComp-5HQ:  ChemComp-5HS:  ChemComp-5GC: 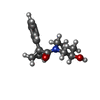 ChemComp-5GJ: 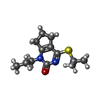 ChemComp-5GL: 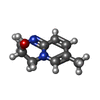 ChemComp-5GQ: 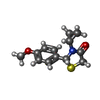 ChemComp-5HJ: 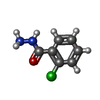 ChemComp-5G1:  ChemComp-601: 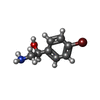 ChemComp-6LY: 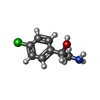 ChemComp-6LE: 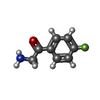 ChemComp-6LZ:  ChemComp-47K: |
| 由来 |
|
 キーワード キーワード | HYDROLASE / fagment screening / inhibition / fragment screening / aspartic protease |
 ムービー
ムービー コントローラー
コントローラー 構造ビューア
構造ビューア 万見文献について
万見文献について



 cryphonectria parasitica (菌類)
cryphonectria parasitica (菌類)