+Search query
-Structure paper
| Title | Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome. |
|---|---|
| Journal, issue, pages | Science, Vol. 376, Issue 6595, Page 844-852, Year 2022 |
| Publish date | May 20, 2022 |
 Authors Authors | Stanislau Yatskevich / Kyle W Muir / Dom Bellini / Ziguo Zhang / Jing Yang / Thomas Tischer / Masa Predin / Tom Dendooven / Stephen H McLaughlin / David Barford /  |
| PubMed Abstract | Kinetochores assemble onto specialized centromeric CENP-A (centromere protein A) nucleosomes (CENP-A) to mediate attachments between chromosomes and the mitotic spindle. We describe cryo-electron ...Kinetochores assemble onto specialized centromeric CENP-A (centromere protein A) nucleosomes (CENP-A) to mediate attachments between chromosomes and the mitotic spindle. We describe cryo-electron microscopy structures of the human inner kinetochore constitutive centromere associated network (CCAN) complex bound to CENP-A reconstituted onto α-satellite DNA. CCAN forms edge-on contacts with CENP-A, and a linker DNA segment of the α-satellite repeat emerges from the fully wrapped end of the nucleosome to thread through the central CENP-LN channel that tightly grips the DNA. The CENP-TWSX histone-fold module further augments DNA binding and partially wraps the linker DNA in a manner reminiscent of canonical nucleosomes. Our study suggests that the topological entrapment of the linker DNA by CCAN provides a robust mechanism by which kinetochores withstand both pushing and pulling forces exerted by the mitotic spindle. |
 External links External links |  Science / Science /  PubMed:35420891 / PubMed:35420891 /  PubMed Central PubMed Central |
| Methods | EM (single particle) / X-ray diffraction |
| Resolution | 2.44 - 12.0 Å |
| Structure data | EMDB-13437, PDB-7pii: EMDB-13473, PDB-7pkn: EMDB-14334, PDB-7r5r: EMDB-14336, PDB-7r5s: EMDB-14341, PDB-7r5v: EMDB-14351, PDB-7ywx: EMDB-14375, PDB-7yyh:  PDB-7pb4:  PDB-7pb8: |
| Chemicals |  ChemComp-HOH: |
| Source |
|
 Keywords Keywords | CELL CYCLE / inner kinetochore / Kinetochore / chromosome segregation / centromere / Chromosome / cell division |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers








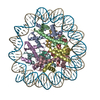








 homo sapiens (human)
homo sapiens (human)