+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the human CCAN deltaCT complex | ||||||||||||
 Map data Map data | 3D consensus refinement | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Kinetochore / chromosome segregation / centromere / CELL CYCLE | ||||||||||||
| Function / homology |  Function and homology information Function and homology information: / Mis6-Sim4 complex / positive regulation of protein localization to kinetochore / kinetochore organization / metaphase chromosome alignment / kinetochore binding / sex differentiation / CENP-A containing chromatin assembly / chordate embryonic development / negative regulation of epithelial cell apoptotic process ...: / Mis6-Sim4 complex / positive regulation of protein localization to kinetochore / kinetochore organization / metaphase chromosome alignment / kinetochore binding / sex differentiation / CENP-A containing chromatin assembly / chordate embryonic development / negative regulation of epithelial cell apoptotic process / kinetochore assembly / inner kinetochore / mitotic sister chromatid segregation / Amplification of signal from unattached kinetochores via a MAD2 inhibitory signal / Deposition of new CENPA-containing nucleosomes at the centromere / Mitotic Prometaphase / EML4 and NUDC in mitotic spindle formation / NRIF signals cell death from the nucleus / Resolution of Sister Chromatid Cohesion / positive regulation of epithelial cell proliferation / mitotic spindle organization / chromosome segregation / RHO GTPases Activate Formins / kinetochore / centriolar satellite / Separation of Sister Chromatids / actin cytoskeleton / chromosome / cell adhesion / nuclear body / cell division / apoptotic process / regulation of DNA-templated transcription / nucleolus / signal transduction / nucleoplasm / membrane / nucleus / cytoplasm / cytosol Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | ||||||||||||
 Authors Authors | Muir KW / Yatskevich S / Bellini D / Barford D | ||||||||||||
| Funding support |  Germany, 3 items Germany, 3 items
| ||||||||||||
 Citation Citation |  Journal: Science / Year: 2022 Journal: Science / Year: 2022Title: Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome. Authors: Stanislau Yatskevich / Kyle W Muir / Dom Bellini / Ziguo Zhang / Jing Yang / Thomas Tischer / Masa Predin / Tom Dendooven / Stephen H McLaughlin / David Barford /  Abstract: Kinetochores assemble onto specialized centromeric CENP-A (centromere protein A) nucleosomes (CENP-A) to mediate attachments between chromosomes and the mitotic spindle. We describe cryo-electron ...Kinetochores assemble onto specialized centromeric CENP-A (centromere protein A) nucleosomes (CENP-A) to mediate attachments between chromosomes and the mitotic spindle. We describe cryo-electron microscopy structures of the human inner kinetochore constitutive centromere associated network (CCAN) complex bound to CENP-A reconstituted onto α-satellite DNA. CCAN forms edge-on contacts with CENP-A, and a linker DNA segment of the α-satellite repeat emerges from the fully wrapped end of the nucleosome to thread through the central CENP-LN channel that tightly grips the DNA. The CENP-TWSX histone-fold module further augments DNA binding and partially wraps the linker DNA in a manner reminiscent of canonical nucleosomes. Our study suggests that the topological entrapment of the linker DNA by CCAN provides a robust mechanism by which kinetochores withstand both pushing and pulling forces exerted by the mitotic spindle. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13473.map.gz emd_13473.map.gz | 15.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13473-v30.xml emd-13473-v30.xml emd-13473.xml emd-13473.xml | 31.8 KB 31.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13473_fsc.xml emd_13473_fsc.xml | 12.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_13473.png emd_13473.png | 52.5 KB | ||
| Filedesc metadata |  emd-13473.cif.gz emd-13473.cif.gz | 8.2 KB | ||
| Others |  emd_13473_additional_1.map.gz emd_13473_additional_1.map.gz emd_13473_additional_2.map.gz emd_13473_additional_2.map.gz emd_13473_additional_3.map.gz emd_13473_additional_3.map.gz emd_13473_additional_4.map.gz emd_13473_additional_4.map.gz | 9.9 MB 9.4 MB 10.8 MB 116.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13473 http://ftp.pdbj.org/pub/emdb/structures/EMD-13473 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13473 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13473 | HTTPS FTP |
-Related structure data
| Related structure data |  7pknMC  7pb4C  7pb8C  7piiC 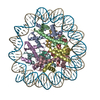 7r5rC  7r5sC  7r5vC  7ywxC  7yyhC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_13473.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13473.map.gz / Format: CCP4 / Size: 149.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D consensus refinement | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: HIKMLN body
| File | emd_13473_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | HIKMLN body | ||||||||||||
| Projections & Slices |
| ||||||||||||
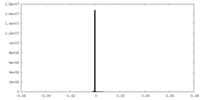
| Density Histograms |
-Additional map: OPQURN body
| File | emd_13473_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | OPQURN body | ||||||||||||
| Projections & Slices |
| ||||||||||||
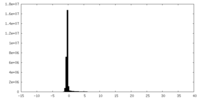
| Density Histograms |
-Additional map: OPQURN focused refinement
| File | emd_13473_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | OPQURN focused refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
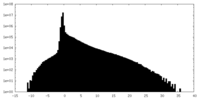
| Density Histograms |
-Additional map: Combined focused map from PHENIX
| File | emd_13473_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Combined focused map from PHENIX | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
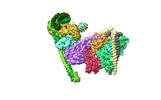
+Entire : Human inner kinetochore CCAN
+Supramolecule #1: Human inner kinetochore CCAN
+Macromolecule #1: Centromere protein H
+Macromolecule #2: Centromere protein I
+Macromolecule #3: Centromere protein K
+Macromolecule #4: Centromere protein L
+Macromolecule #5: Centromere protein M
+Macromolecule #6: Centromere protein N
+Macromolecule #7: Centromere protein O
+Macromolecule #8: Centromere protein P
+Macromolecule #9: Centromere protein Q
+Macromolecule #10: Centromere protein U
+Macromolecule #11: Centromere protein R
+Macromolecule #12: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)















































 Trichoplusia ni (cabbage looper)
Trichoplusia ni (cabbage looper)

