+Search query
-Structure paper
| Title | Structural basis for the inhibition of the eukaryotic ribosome. |
|---|---|
| Journal, issue, pages | Nature, Vol. 513, Page 517-522, Year 2014 |
| Publish date | Jul 22, 2014 (structure data deposition date) |
 Authors Authors | Garreau de Loubresse, N. / Prokhorova, I. / Holtkamp, W. / Rodnina, M.V. / Yusupova, G. / Yusupov, M. |
 External links External links |  Nature / Nature /  PubMed:25209664 PubMed:25209664 |
| Methods | X-ray diffraction |
| Resolution | 2.801 - 3.6 Å |
| Structure data |  PDB-4u3m:  PDB-4u3n:  PDB-4u3u:  PDB-4u4n:  PDB-4u4o:  PDB-4u4q:  PDB-4u4r:  PDB-4u4u:  PDB-4u4y:  PDB-4u4z:  PDB-4u50:  PDB-4u51:  PDB-4u52:  PDB-4u53:  PDB-4u55:  PDB-4u56:  PDB-4u6f: |
| Chemicals |  ChemComp-MG: 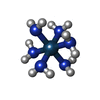 ChemComp-OHX:  ChemComp-ZN:  ChemComp-ANM: 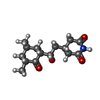 ChemComp-3HE:  ChemComp-EDE:  ChemComp-GET:  ChemComp-HMT: 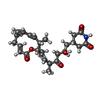 ChemComp-3H3:  ChemComp-3KD:  ChemComp-PCY:  ChemComp-3K5:  ChemComp-3L2:  ChemComp-3KF: 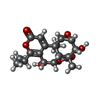 ChemComp-3J2:  ChemComp-3J6:  ChemComp-3K8: 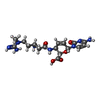 ChemComp-BLS: 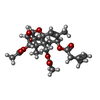 ChemComp-ZBA: |
| Source |
|
 Keywords Keywords | RIBOSOME / translation / 40S / 60S / 80S / eukaryote / RNA-protein complex / inhibitor / antibiotic |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers




